The if a: else b statement is one of the most common statements in Python programming. By nesting and combining such statements, we can build a so-called decision tree. This is similar to an old fashioned flowchart, although flowcharts also allow loops. The application of decision trees in machine learning is called decision tree learning. The end nodes of the trees in decision tree learning, also known as leaves, contain the class labels of a classification problem. Each non-leaf node is associated with a Boolean condition involving feature values.
Decision trees can be used to deduce relatively simple rules. Being able to produce such results is, of course, a huge advantage. However, you have to wonder how good these rules are. If we add new data, would we get the same rules?
If one decision tree is good, a whole forest should be even better. Multiple trees should reduce the chance of overfitting. However, as in a real forest, we don't want only one type of tree. Obviously, we would have to average or decide by majority voting what the appropriate result should be.
In this recipe, we will apply the random forest algorithm invented by Leo Breiman and Adele Cutler. The "random" in the name refers to randomly selecting features from the data. We use all the data but not in the same decision tree.
Random forests also apply bagging (bootstrap aggregating), which we will discuss in the Bagging to improve results recipe. The bagging of decision trees consists of the following steps:
- Sample training examples with replacement and assign them to a tree.
- Train the trees on their assigned data.
We can determine the correct number of trees by cross-validation or by plotting the test and train error against the number of trees.
The code is in the random_forest.ipynb file in this book's code bundle:
- The imports are as follows:
import dautil as dl from sklearn.grid_search import GridSearchCV from sklearn.ensemble import RandomForestClassifier import ch9util import numpy as np from IPython.display import HTML
- Load the data and do a prediction as follows:
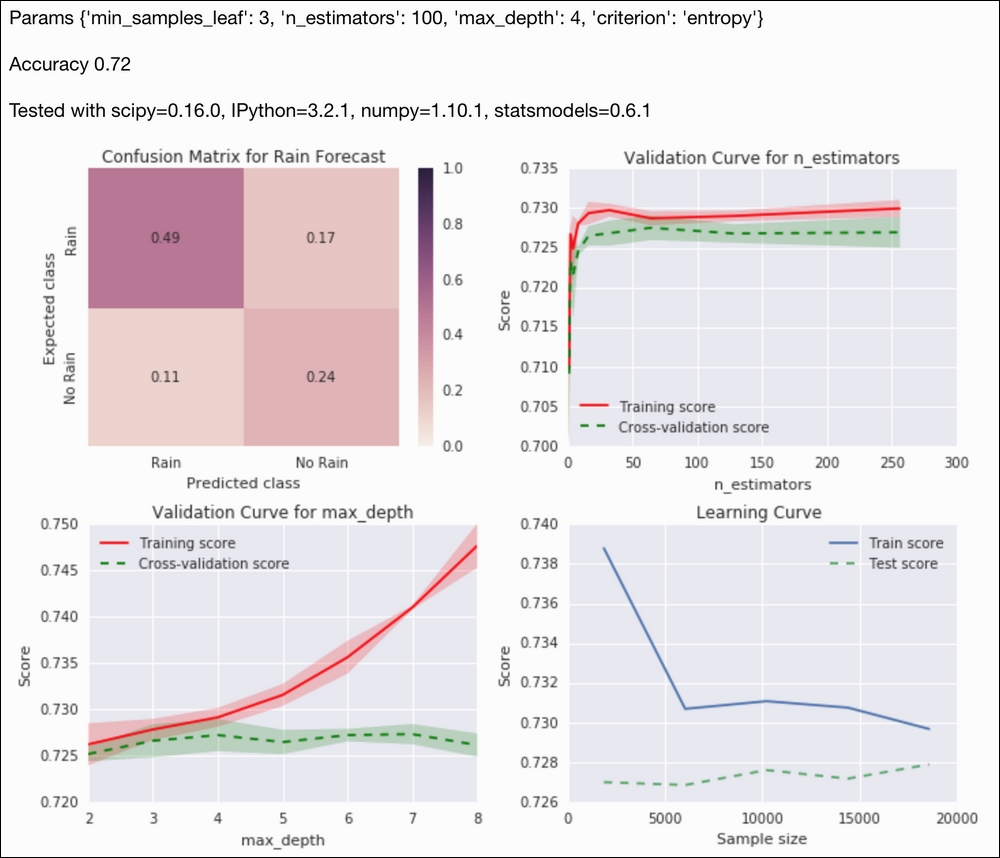
X_train, X_test, y_train, y_test = ch9util.rain_split() clf = RandomForestClassifier(random_state=44) params = { 'max_depth': [2, 4], 'min_samples_leaf': [1, 3], 'criterion': ['gini', 'entropy'], 'n_estimators': [100, 200] } rfc = GridSearchCV(estimator=RandomForestClassifier(), param_grid=params, cv=5, n_jobs=-1) rfc.fit(X_train, y_train) preds = rfc.predict(X_test) - Plot the rain forecast confusion matrix as follows:
sp = dl.plotting.Subplotter(2, 2, context) html = ch9util.report_rain(preds, y_test, rfc.best_params_, sp.ax)
- Plot a validation curve for a range of forest sizes:
ntrees = 2 ** np.arange(9) ch9util.plot_validation(sp.next_ax(), rfc.best_estimator_, X_train, y_train, 'n_estimators', ntrees) - Plot a validation curve for a range of depths:
depths = np.arange(2, 9) ch9util.plot_validation(sp.next_ax(), rfc.best_estimator_, X_train, y_train, 'max_depth', depths) - Plot the learning curve of the best estimator:
ch9util.plot_learn_curve(sp.next_ax(), rfc.best_estimator_, X_train,y_train) HTML(html + sp.exit())
Refer to the following screenshot for the end result:
Random forests classification is considered such a versatile algorithm that we can use it for almost any classification task. Genetic algorithms and genetic programming can do a grid search or optimization in general.
We can consider a program to be a sequence of operators and operands that generates a result. This is a very simplified model of programming, of course. However, in such a model, it is possible to evolve programs using natural selection modeled after biological theories. A genetic program is self-modifying with huge adaptability, but we get a lower level of determinism.
The TPOT project is an attempt to evolve machine learning pipelines (currently uses a small number of classifiers including random forests). I forked TPOT 0.1.3 on GitHub and made some changes. TPOT uses deap for the genetic programming parts, which you can install as follows:
$ pip install deap
I tested the code with deap 1.0.2. Install my changes under tag r1 as follows:
$ git clone [email protected]:ivanidris/tpot.git $ cd tpot $ git checkout r1 $ python setup.py install
You can also get the code from https://github.com/ivanidris/tpot/releases/tag/r1. The following code from the rain_pot.py file in this book's code bundle demonstrates how to fit and score rain predictions with TPOT:
import ch9util from tpot import TPOT X_train, X_test, y_train, y_test = ch9util.rain_split() tpot = TPOT(generations=7, population_size=110, verbosity=2) tpot.fit(X_train, y_train) print(tpot.score(X_train, y_train, X_test, y_test))
- The Wikipedia page about random forests at https://en.wikipedia.org/wiki/Random_forest (retrieved November 2015)
- The documentation for the
RandomForestClassifierclass at http://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html (retrieved November 2015)
