19
Image Segmentation of Neuronal Cell with Ensemble Unet Architecture
Kirtan Kanani1, Aditya K. Gupta1, Ankit Kumar Nikum1⋆, Prashant Gupta2 and Dharmik Raval1
1Department of Mechanical, Sardar Vallabhbhai National Institute of Technology, Surat, Gujarat, India
2Licious, Banglore, India
Abstract
Medical image segmentation consists of heterogeneous pixel intensities, noisy/ ill-defined borders, and high variability, which are significant technical obstacles for segmentation. Also, generally the requirement of annotated samples by the networks is significantly large to achieve high accuracy. Gathering this dataset for the particular application and annotating new images is both time-consuming and costly. Unet solves this problem by not requiring vast datasets for picture segmentation. The present work describes the use of a network that depends on augmentation of the existing annotated dataset to make better use of these examples and a comparison of encoder accuracy on Unet is presented. The encoder principal function is to reduce image dimensionality while keeping as much information as possible. EfficientNets tackles both of these issues and utilizing it as an encoder of Unet can further enhance its accuracy. The test dataset highest F1-Score and IoU were 0.7655 and 0.6201 on neuronal data values, respectively. It outperforms Inception and ResNet encoder networks with considerably more parameters and a higher inference time.
Keywords: Image segmentation, computer vision, deep learning, neural cell
19.1 Introduction
Neurodegenerative illnesses cover a diverse spectrum of ailments resulting from progressive damage to cells and nervous system connections. These illnesses eventually lead to diseases such as Alzheimer’s and Parkinson’s disease [1], which are among the six highest causes of death. According to the WHO report [2] on neurological disorders, neurological illnesses account for 12% of all fatalities worldwide. Figure 19.1 shows the architecture of the modified U-Net model used for training. The goal of finding treatments and solutions for neurodegenerative illnesses is becoming more urgent with the increase in population. In 2015, neurological illnesses caused 250.7 million disability-adjusted life years (DALYs) and 9.4 million deaths, accounting for 10.2% of global DALYs and 16.8% of global deaths [3]. Our work is focused on assisting doctors in determining the effects of medical treatments in neuron cells using machine learning and computer vision.
For biomedical image analysis, the segmentation of 2D pictures is a critical problem [4]. We have proposed an Unet based neural network that upgrades the FCN model for biomedical application, i.e., RIC Unet, for nuclei segmentation. Following Unet architecture, RIC-Unet uses a channel attention mechanism, residual blocks, and multi-scale techniques for segmentation of nuclei accurately. This network is superior to other techniques in terms of analysing indicators and cost-effectiveness in assisting doctors in diagnosing the details of these histological images.

Figure 19.1 Architecture of Unet with EfficentNet as encoder. The decoder is similar to the original Unet model but with cropping and contamination from various encoder elements.
In many medical picture segmentation issues U-Net provides a state-of-the-art performance. U-Net with residual blocks or blocks with dense connections, Attention U-Net, and recurrent residual convolutional U-Net are a few examples of U-Net adaptations. All of these adaptations have some changes in common such as the ways in which the path of information flow is altered and an encoder-decoder structure with skip connections. In this work, the author was inspired by the success of ResNet and R2-UNet [5] employing modified residual blocks. Here, the two convolutional layers in one block are sharing the same weights [5].
For the segmentation of cell nuclei images [6], the intersection over union (IoU) is used as a quality matrix for increasing the performance of Unet, ResUNet, and DeepLab. The proposed model has a mean IoU value of 0.9005 for validation dataset.
19.2 Methods
The Unet framework mainly inspired our architecture. We utilized EfficientNet-B0 as an encoding part. Like Unet, encoder branch and decoder branch features are concatenated. We chose EfficientNet as the encoder because it proposes an effective compound scaling strategy that effectively scales up a baseline ConvNet to any target resource limitations in a more principled manner while retaining model efficiency.
The ImageNet dataset containing color images was used for training the original EfficientNet. The images had a resolution of 224×224. In the present study, a pre-trained EfficientNet model was implemented as an encoder part of Unet to reduce the training time. The EfficientNet-B0 with low parameter quantity was applied owing to limited complexity and an easily identifiable nature of the neuronal images.
19.3 Dataset
The dataset consists of 606 grayscale images with an image size of 520 × 704 pixels. We chose 364 photos at random for training, 61 for validation, and 60 for testing. Image count of ‘shsy5y’, ‘Astro’, ‘Cort’ was 155, 131, and 320, respectively. The neuroblastoma cell line Cort consistently has the lowest precision scores of the three cancer cell lines. This could be because neuronal cells have a distinct, uneven, and concave look, making them difficult to segment using standard mask heads.
19.4 Implementation Details
Data augmentation is being performed which includes random cropping, random horizontal/vertical flipping, 35° rotation, and further enhanced accuracy normalization is used on the training images with various schemes. In this study, the call back function was implemented, which monetarized the validation loss for 150 training epochs. If there is not a significant change in this value, then training is terminated. The training and testing images have a resolution of 256 × 256 pixels. We used binary cross-entropy (BCE) with sigmoid as a loss function and Adam as an optimizer. The initial learning rate was set to 1e-4 to optimize the model weights and PyTorch on Tesla P100-PCIE-16GB was used to implement the model.
19.5 Evaluation Metrics
Average Precision (AP) [7] is used as a metric to evaluate both instances’ segmentation performance. For instance, in segmentation, we employ APmask in Equation 19.1 to represent the AP at the mask and the IoU between the ground truth mask and each predicted segmentation mask.
19.6 Result
For this work of semantic segmentation, we used a variety of designs and the results are given in Table 19.1. Our investigation began with the DeepLabV3+ framework. For extraction of features, we initially utilized EfficientNet-B0 as an encoder. We experimented with several encoder backbones such as EfficientNet-B0 [8], ResNet-50 [9], InceptionResNetV2 [10], and EfficientNet-B5 with the renowned Unet decoder. From Table 19.1, it can be shown that the Unet decoder outperforms the DeepLabV3+ decoder. That is because Unet now has additional low-level encoder characteristics that are essential in analyzing complicated scenes with multiple dense objects.
Table 19.1 Result comparison in terms of mean intersection over union (IoU), F1-score, and accuracy on the neuronal dataset.
| Model | Fl-score | IoU | Accuracy |
|---|---|---|---|
| Unet with EfficientNet-B0 as encoder (binary cross entropy logits) | 0.7286 | 0.573 | 0.9466 |
| Unet with ResNet-50 as encoder (binary cross entropy logits) | 0.7053 | 0.5447 | 0.9254 |
| Unet with Inception-ResNet-v2 as encoder (binary cross entropy logits) | 0.7264 | 0.5704 | 0.9550 |
| Unet with EfficientNet-B0 as encoder (dice loss) | 0.7655 | 0.6201 | 0.9493 |
| PSPNet with EfficientNet-B2 as encoder (binary cross entropy logits) | 0.6075 | 0.4362 | 0.9321 |
| PSPNet with InceptionNet as encoder (binary cross entropy logits) | 0.6579 | 0.4902 | 0.9311 |
| DeeplabV3+ with EfficientNet-B5 as encoder (binary cross entropy logits) | 0.7089 | 0.5490 | 0.9351 |
| Unet with Inception-ResNet-v2 as encoder (dice loss) | 0.7283 | 0.5727 | 0.9166 |
| DeeplabV3+ with EfficientNet-B5 as encoder (dice loss) | 0.7265 | 0.5705 | 0.9351 |
We further analyze the class-wise segmentation performance of the best-performing model on the three classes. We obtain class-wise IoU on the validation dataset, shown in Table 19.2. It can be observed that the Cort cell has the lowest IoU. The Cort cell image contains different geometry types compared to Astro and sphy5y. Besides the circular geometry of Cort cells, thread-like structures are also present in the image, which is similar to Astro and shyishy5 geometry. This makes model prediction clouded. The model’s accuracy is further hampered by a small dataset. The ground truth segmentation maps of a few images, as well as their anticipated segmentation maps, are shown in Figure 19.2.
Table 19.2 Class wise IoU on neuronal dataset with efficient Unet as model.
| Neuronal cells | shsy5y | Astro | Cort |
|---|---|---|---|
| Mean IoU | 0.4580 | 0.5474 | 0.2948 |
The first column displays the input photos for a single cell. The ground truth and anticipated segmentation maps are shown in the second and third columns, respectively. The cell type is in the order Astro, Cort, Shsy5y from top to bottom.
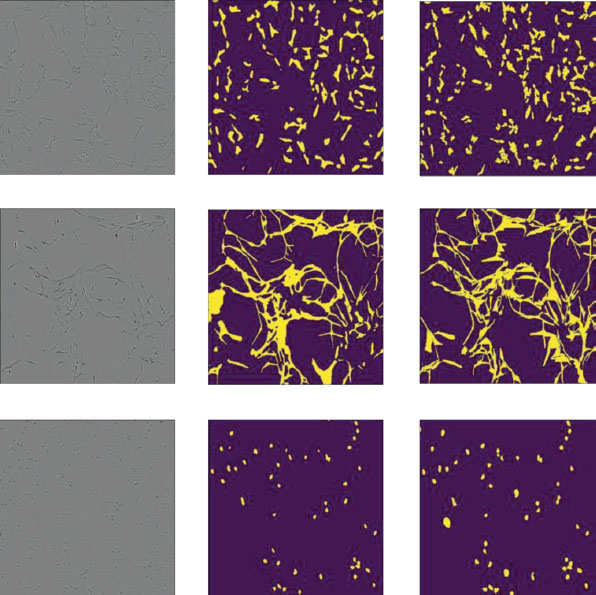
Figure 19.2 Result of semantic segmentation on neuronal validation dataset with efficient Unet as architecture.
19.7 Conclusion
This study employs multiple kinds of encoders for image segmentation and is based on Unet architecture. It provides a comprehensive procedure that includes selecting a data set, acquiring a training set, training a deep convolutional neural network, and segmenting cell images using the convolution neural network. Finally, following cell image segmentation, the Unet with various encoders was utilized to generate the resultant image, and a basic analysis of the acquired values was performed. This technique achieves high-precision semantic segmentation of shsy5y cells and Astro pictures compared to traditional image segmentation.
References
- 1. S. Przedborski, M. Vila, and V. Jackson-Lewis, ‘Series Introduction: Neurodegeneration: What is it and where are we?’, J. Clin. Invest., vol. 111, no. 1, p. 3, Jan. 2003, doi: 10.1172/JCI17522.
- 2. W. H. Organization, ‘Neurological disorders: public health challenges’, World Health Organization, 2006.
- 3. V. L. Feigin et al., ‘Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016’, Lancet Neurol., vol. 18, no. 5, pp. 459–480, May 2019, doi: 10.1016/S1474-4422(18)30499-X.
- 4. Z. Zeng, W. Xie, Y. Zhang, and Y. Lu, ‘RIC-Unet: An Improved Neural Network Based on Unet for Nuclei Segmentation in Histology Images’, IEEE Access, vol. 7, pp. 21420–21428, 2019, doi: 10.1109/ACCESS.2019. 2896920.
- 5. J. Zhuang, ‘LadderNet: Multi-path networks based on U-Net for medical image segmentation’, Oct. 2018, Accessed: Dec. 13, 2021. [Online]. Available: https://arxiv.org/abs/1810.07810v4.
- 6. C. A. R. Goyzueta, J. E. C. De la Cruz, and W. A. M. Machaca, ‘Integration of U-Net, ResU-Net and DeepLab Architectures with Intersection Over Union metric for Cells Nuclei Image Segmentation’, pp. 1–4, Nov. 2021, doi: 10.1109/EIRCON52903.2021.9613150.
- 7. M. Everingham, L. Van Gool, C. K. I. Williams, J. Winn, and A. Zisserman, ‘The pascal visual object classes (VOC) challenge’, Int. J. Comput. Vis., vol. 88, no. 2, pp. 303–338, Sep. 2010, doi: 10.1007/s11263-009-0275-4.
- 8. M. Tan and Q. V. Le, ‘EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks’, 36th Int. Conf. Mach. Learn. ICML 2019, vol. 2019-June, pp. 10691–10700, May 2019, Accessed: Dec. 13, 2021. [Online]. Available: https://arxiv.org/abs/1905.11946v5.
- 9. K. He, X. Zhang, S. Ren, and J. Sun, ‘Deep Residual Learning for Image Recognition’, Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2016-December, pp. 770–778, Dec. 2015, doi: 10.1109/CVPR.2016.90.
- 10. C. Szegedy, S. Ioffe, V. Vanhoucke, and A. A. Alemi, ‘Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning’, 31st AAAI Conf. Artif. Intell. AAAI 2017, pp. 4278–4284, Feb. 2016, Accessed: Dec. 15, 2021. [Online]. Available: https://arxiv.org/abs/1602.07261v2.
Note
- ⋆ Corresponding author: [email protected]
