Perform the following steps in R:
- Find the minimum cross-validation error of the classification tree model:
> install.packages("mice")
> install.packages("randomForest")
> install.packages("VIM")
> t = data.frame(x=c(1:100), y=c(1:100))
> t$x[sample(1:100,10)]=NA
> t$y[sample(1:100,20)]=NA
> aggr(t)
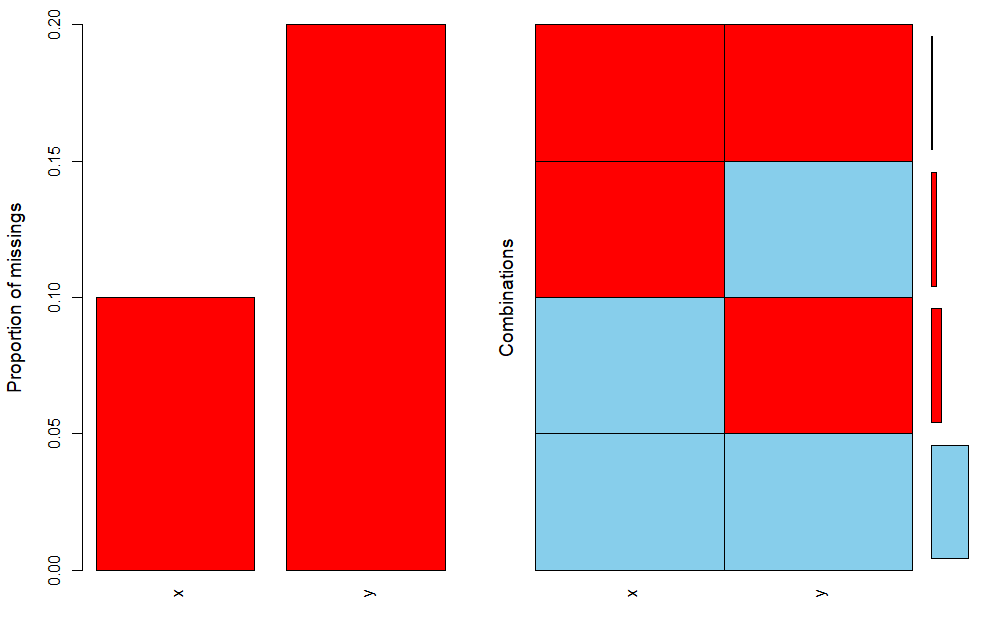
- Tweaking the aggr function by adding prop and number will produce a slightly different output showing numbers for combinations also:
> aggr(t, prop=T, numbers=T)

> matrixplot(t)

> md.pattern(t)
Output
x y
71 1 1 0
9 0 1 1
19 1 0 1
1 0 0 2
10 20 30
> m <- mice(t, method="rf")
> m
Output
iter imp variable
1 1 x
> mc <- complete(m)
> anyNA(mc)
Output
[1] False
