Chapter 11: Deep learning-based detection and classification of adenocarcinoma cell nuclei
Abstract
Nowadays, clinical practice uses digital pathology for examining digitized microscopic images to identify diseases like cancers. The main challenge in examining microscopic pictures is the need to dissect every single individual cell for precise analysis because identification of cancerous diseases depends emphatically on cell-level data. Due to this reason, the detection of cells is a significant point in medical image examination, and it is regularly the essential prerequisite for disease classification techniques. Cell detection and then classification is a problematic issue because of diverse heterogeneity in the characteristics of the cells. Deep learning methodologies have appeared to deliver empowering results on analyzing digital pathology pictures. This chapter introduces an approach by using region-based convolution neural networks for locating the cell nuclei. The region-based convolution neural network estimates the probability of a pixel belonging to the core of the cell nuclei. Pixels with maximum probability indicate the location of the core of the cell nucleus. After finding the cells, they are classified as healthy or malicious cells by training a deep convolution neural network. The proposed approach for cell detection and classification is tested with the adenocarcinoma dataset. Cell image analysis based on deep learning techniques shows good results in both the identification and classification of the cell nucleus.
Keywords
1. Introduction
2. Basics of a convolution neural network
2.1. Convolution layer
- • Filters: The number of filters.
- • Kernel_size: A number specifying both the height and width of the (square) convolution window. Some additional optional arguments might be tuned.
- • Strides: The stride of the convolution. If the user does not specify anything, it is set to 1.
- • Padding: This is either valid or the same. If the user does not specify anything, the padding is set to valid.
- • Activation: This is typically ReLu. If the user does not specify anything, no activation is applied. It is strongly advised to add a ReLU activation function to every convolution layer in the networks.
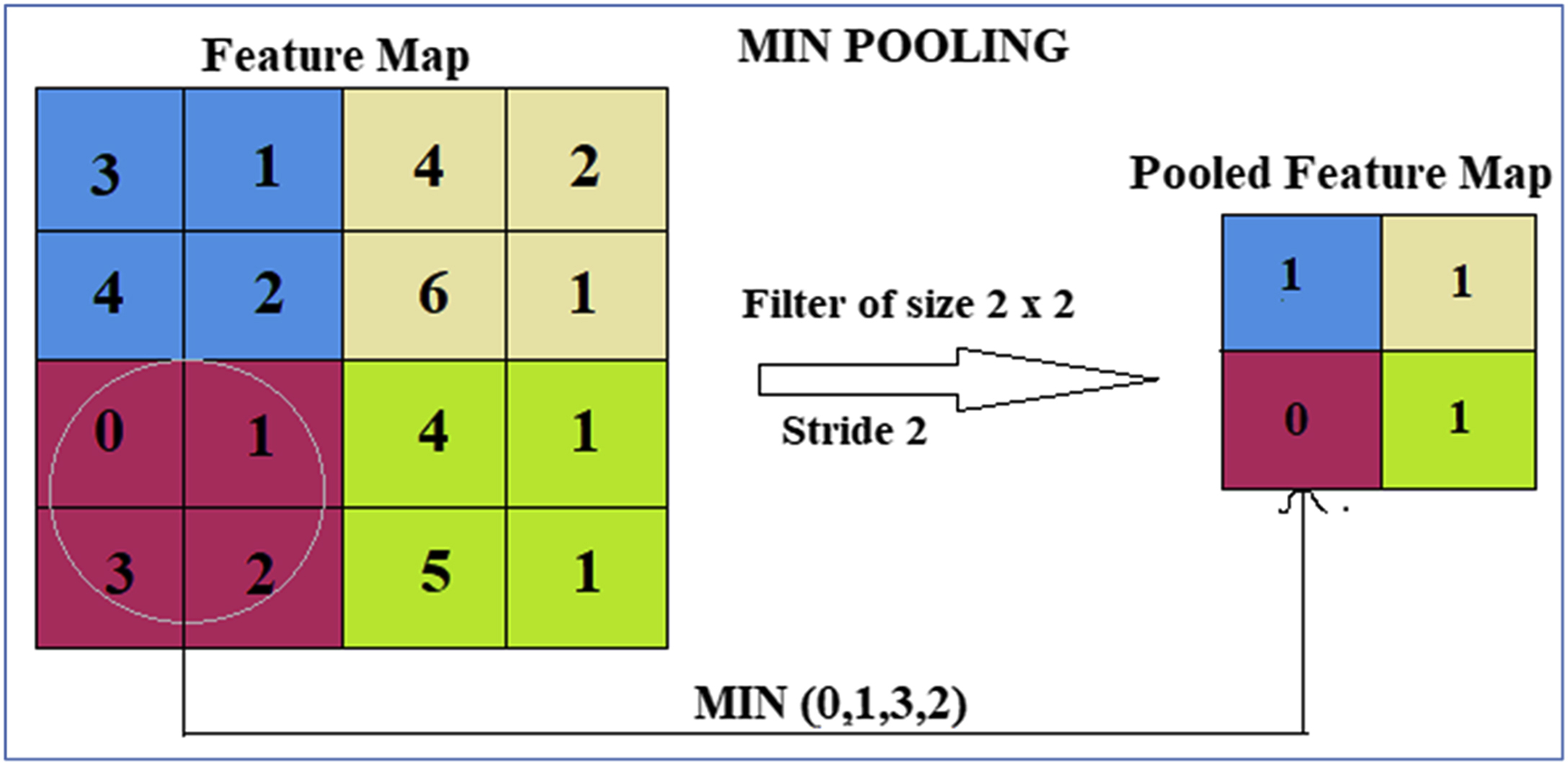
2.2. Pooling layer
2.3. Fully connected layer
3. Literature review
4. Proposed system architecture and methodology
Table 11.1
| References | Application | Architecture used | Learning type |
|---|---|---|---|
| Hao Chen [17] | Cell detection | Deep neural network | Design and training |
| Cires et al. [20] | Cell detection | CNN | Design and training |
| Janowczyk and Madabhushi [22] | Identifying lymphocytes in breast cancer images | CNN | Transfer learning |
| Khoshdeli et al. [23] | Checking nuclei in hematoxylin and eosin marked metaphors | Deep neural network | Design and training |
| Y. Xue [24] | Cell checking and recognition | CNN | Design and training |
| Henning Höfener et al. [25] | Identification and classification of nuclei | CNN | Design and training |
| Chowdhurya et al. [26] | Grade classification in colon cancer | CNN | Transfer learning |
| Kashif et al. [27] | Detection of tumor cells | CNN | Design and training |
| Xu et al. [28] | Feature learning with minimum manual annotation | CNN | Design and training |
| Haj-Hassan et al. [29] | Classification of cells corresponding to colorectal cancer | CNN | Design and training |

- Step 1: Load the image dataset
- Step 2: Adjust the training and test sets
- Step 3: Detect the cells with Faster R-CNN
- Step 4: Prepare training images and test image sets
- Step 5: Load the pretrained ResNet-101 model
- Step 6: Train the ResNet-101 with the training data
- Step 7: Evaluate the classifier
- Step 8: Apply the trained classifier to test images
- Step 9: Evaluate the accuracy
4.1. Cell detection using faster R-CNN

- Step 1: Pass an input image to the convolution net of Faster R-CNN, which retrieves the feature map in the given input.
- Step 2: The identified feature map is the given to the region proposal network (RPN) of Faster R-CNN. The RPN receives the object proposals.
- Step 3: The identified object proposals are given to the region of interest (RoI) pooling layer to convert the object proposals of varying sizes into a uniform size.
- Step 4: The uniformly converted object proposals are passed onto the fully connected layers of Faster R-CNN. The fully connected layers place bounding boxes if the object proposal contains a cell that is different from the background.
4.2. Cell classification with ResNet-101
5. Experimentation
5.1. Dataset
5.2. Discussion on results