Chapter 14. Curating cancer research data with Tika
For more than 10 years, the goal of the National Cancer Institute’s (NCI) Early Detection Research Network (EDRN) program has been to accelerate research into the identification and detection of cancer biomarkers, early indicators of disease.
Over the last three years, Tika has been assisting in that fight. Tika helps capture and curate mass spectrometry files, CSV files, PDF files, SAS files, and other data sets produced by lab instruments, analysis programs, and by EDRN investigators for further scientific research and role-based dissemination to the broader community. In this chapter, we’ll explain how Tika has been used in EDRN’s Catalog and Archive Service, or eCAS. Let’s hop to it.
14.1. The NCI Early Detection Research Network
The National Cancer Institute’s EDRN project has focused on the identification of cancer biomarkers for over a decade. Biomarkers are indicators of early onset of disease, and identifying accurate and precise methods for their detection is one of the EDRN’s primary goals.
As part of the biomarker discovery process, EDRN collects many different types of data, ranging from information about biomarkers themselves (names, aliases, related organs, and so forth), to instrument-produced raw science data files, study protocols, and specimen information. We’ll talk about these types in the next section.
14.1.1. The EDRN data model
EDRN’s (simplified) data model is shown in figure 14.1. We’ll discuss the relationships between each of the major data types and the relationships of these types to Tika. As we’ll detail, Tika helps identify, sort, and extract metadata from EDRN’s science data sets, shown in the middle of the figure.
Figure 14.1. Simplified view of the EDRN data model, showing the relationship between protocols, specimens, science data sets, biomarkers, and instruments

In EDRN, research information about biomarkers, science data sets (collections of files produced by an instrument), and specimens (taken during some associated EDRN study), are all guided by common study information called a protocol. Protocols are led by principal investigators, potentially from multiple EDRN participating sites.
With that in mind, let’s restrict our focus to the science data set portion of the EDRN data model, shown conveniently in the middle portion of figure 14.1. There are myriad science instruments in the EDRN, ranging from high-end mass spectrometers, to biospecimen catalog tools, to low-end microscopes. Each of these instruments produces data associated with some EDRN biomarker, and some EDRN study, and is of interest to the broader program.
This is where Tika comes in. The process of collecting, curating, annotating, and making these data sets available to the rest of the EDRN program is called scientific data curation, and we’ll explain it in more detail in the next section.
14.1.2. Scientific data curation
More and more, the community at large is seeing the importance of data curation. Data curation involves the careful preparation of data for cataloging, archiving, and eventual dissemination to the external community. Just like librarians curate books that codify our collective knowledge, data curators (and the associated tools they use) do the same thing for data.
One of the key goals of the EDRN is to capture rich metadata and data that conforms to the EDRN data model described in figure 14.1. Specifically, EDRN investigators, program managers, and users want to query the system and determine the current state of progress on a particular biomarker or set of biomarkers. This includes answering questions such as the following:
- What publications have been generated for a particular EDRN biomarker?
- What associated science data files are available to help reproduce the experiments and results described in those publications?
- What protocols being studied have produced the most biomarkers?
In order to answer these questions, scientific data curation within the EDRN requires the creation of linked data, a term used to reference richly curated and annotated information captured by an information system.
The EDRN curation process is led by the EDRN biocurator, which sits between computer scientists and cancer researchers helping to capture needed information in a way that leverages the underlying technology but is also scientifically meaningful. Biocurators require the ability to perform both “lights-out” ingestion of day-to-day information, as well as carefully review and annotate that data with additional publication information, study and protocol information, and anything else that the biocurator deems useful to link to the existing EDRN datasets.
A key enabling technology in the EDRN curation process is Tika. Let’s find out in more detail how Tika enables this functionality.
14.2. Integrating Tika
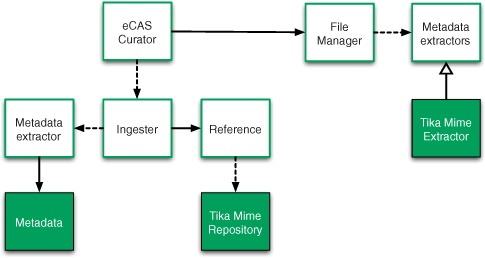
Within EDRN, we’ve built a system called eCAS, which stands for the EDRN Catalog and Archive Service. eCAS provides data curation services for the EDRN and builds upon Apache Tika (along with a number of other technologies, including the Apache OODT framework that we’ll mention in an upcoming sidebar), as figure 14.2 and the ensuing discussion will show. The eCAS system is responsible for providing the necessary software to allow rich metadata and information to be extracted and captured, and to be made available to the rest of the EDRN.
Figure 14.2. EDRN’s eCAS architecture. The components on the left side of the diagram use Tika to prepare data for ingestion into a file manager. The components on the right side of the diagram use Tika to classify incoming files. The components that are directly implemented by Tika are shaded in grey.
It’s Not Just for Curating Nasa Data; It Works for Biomarkers Too! Apache OODT (http://oodt.apache.org) is the first NASA project to be contributed to and hosted at the Apache Software Foundation. OODT includes a number of components that deal with file management, workflow management, resource management, and crawling and archiving sets of files and metadata. It’s the perfect bootstrapping framework to feed information to Tika. It’s a vast framework though, so much so that we can’t go into detail on it here. A careful description of OODT in itself is probably the topic of a forthcoming book.
The first step in EDRN data curation is the extraction of metadata before ingestion.
14.2.1. Metadata extraction
EDRN curators need the ability to perform metadata extraction at various stages throughout the data lifecycle. Initially, data files may be delivered in two methodologies: offline or online. Offline methods include hard disks, CD/DVD ROMs, and so forth. Online methods include electronic protocols such as WebDAV and FTP. Once the data arrives at the EDRN, the eCAS Curator provides a view into the staging area, as shown in the leftmost portion of figure 14.3.
Figure 14.3. The EDRN curation cockpit. The left side of the web application focuses on the staging area, allowing a curator to perform metadata extraction and manipulation. The right side of the webapp deals with data curation and ingestion in the File Manager component, allowing for classification of different file types. Most of the underlying extraction and classification functionality is driven by Tika.

One of the most important parts about data curation in EDRN is capturing rich metadata that can be used to later discover what information is available on a particular EDRN biomarker. For this purpose, we’ve constructed a curation system called eCAS Curator, a webapp (we’ll show you what it looks like later in the chapter) that orchestrates the curation process as shown in figure 14.2. eCAS Curator invokes an Ingester component that’s responsible for extraction of metadata from incoming EDRN files delivered from external sites and investigators. The Ingester is also responsible for generation of file location information, called references.
Tika assists in the eCAS curation process in two ways. First, Tika helps extract additional client-side metadata on a per-file basis, including file naming convention metadata (recall chapter 8) and other information that becomes useful later during the curation process. This is depicted in the left side of figures 14.2 and 14.3 with the Metadata class, extracted from files by Tika. After assisting with metadata extraction, Tika is also used to classify files to be ingested. This is performed using the Tika Mime Repository shown in the middle of figure 14.2. The Reference class uses Tika to determine the underlying MIME type classification for files collected during the curation process. A snippet of code extracted from Apache OODT’s Reference class (part of the File Manager module) is shown next.
Listing 14.1. Detecting file types prior to ingestion in the eCAS system

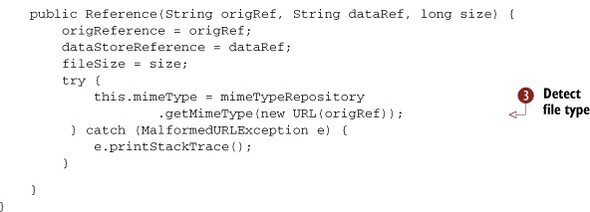
The code in listing 14.1 illustrates that all eCAS Reference class implementations store a Tika MimeType used to capture the MIME type, along with its fully qualified name (recall chapter 4), illustrated in
![]() . This information is detected when the Reference class constructor is called in
. This information is detected when the Reference class constructor is called in
![]() . The Reference class tries to load a user-specified Tika MIME type configuration file in
. The Reference class tries to load a user-specified Tika MIME type configuration file in
![]() and if it can’t be found for whatever reason, then Tika’s default MIME type repo is utilized. Though OODT provides other constructors
for the Reference class, this auto-detect constructor is leveraged during the EDRN ingestion process.
and if it can’t be found for whatever reason, then Tika’s default MIME type repo is utilized. Though OODT provides other constructors
for the Reference class, this auto-detect constructor is leveraged during the EDRN ingestion process.
The middle of figure 14.2 depicts the File Manager component, responsible for taking the extracted Metadata and Reference information and cataloging and archiving it. Part of this process involves the creation of derived metadata (final file locations, data versions, file received time, and so on) that’s best generated on the server side. Another part of this process involves determining and classifying data sets, a process that involves Tika and that we’ll describe in the next section.
14.2.2. MIME type identification and classification
Tika helps the EDRN eCAS system classify and organize collected data files based on more granular MIME types as well as file collection types called data sets in EDRN terminology.
To compute server-side Metadata, Tika is also leveraged as shown in the right side of figure 14.2 with the Tika Mime Extractor component. This server-side metadata extractor makes the extracted MIME type information searchable in the underlying File Manager catalog, breaking the MIME type down into its primary (text in text/html) and subtype (html in text/html) in addition to cataloging its full form (text/html). This functionality is shown in the following listing.
Listing 14.2. Making extracted MIME information available for search and retrieval

Appending the full, primary, and subtypes to the extractMet variable as shown in listing 14.2 allows this information to flow through to the underlying File Manager catalog used by eCAS. Also, allowing the MIME type to be broken up into subparts allows EDRN users to search for specific
kinds of primary files. For example, you could search for all text files associated with a particular biomarker, while retaining
the flexibility to drill down into specifics such as subfile categories and data set types. To perform this functionality,
during extraction, the Product class and its first Reference instance are looked up as shown in
![]() . After locating the first ProductReference instance, the MIME type information is added as shown in
. After locating the first ProductReference instance, the MIME type information is added as shown in
![]() .
.
Besides driving search and display of metadata, the MIME information extracted by Tika is also used to display download links to EDRN data files, and to determine whether the underlying file is capable of being displayed by the browser (in case the primary type is image), versus directly streaming the data file back to the user from the browser link.
At this point, we’ve covered all of the primary areas where Tika has helped out in the EDRN. Let’s review the chapter.
14.3. Summary
We explained the US National Cancer Institute’s Early Detection Research Network (EDRN) project, and how the goal of the project is to develop accurate and precise means of identifying cancer biomarkers, early indicators of disease.
Part of the identification process involves understanding the types of data made available within the EDRN, ranging from specimens collected during a protocol or study, data sets associated with instruments that operate on those specimens, biomarker information providing an up-to-date look into the research progress within EDRN, and PI (principal investigator) and investigator information used to track who’s researching what and how far they are.
The system within EDRN that allows for the capture, preparation, and dissemination of EDRN science data files is the EDRN Catalog and Archive Service. eCAS heavily leverages Tika to provide needed functionality, including file type classification, metadata extraction (both client- and server-side), search, and data download.
We explained each of these areas of eCAS in detail and described where and how Tika was used to implement key eCAS functionality.
We’ve made it to the top of the hill. Only one more chapter to go, and it’s fitting that it begins where the book itself began—with a classic search engine example!
