Building disease and target knowledge with Semantic MediaWiki
Abstract:
The efficient flow of both formal and tacit knowledge is critical in the new era of information-powered pharmaceutical discovery. Yet, one of the major inhibitors of this is the constant flux within the industry, driven by rapidly evolving business models, mergers and collaborations. A continued stream of new employees and external partners brings a need to not only manage the new information they generate, but to find and exploit existing company results and reports. The ability to synthesise this vast information ‘substrate’ into actionable intelligence is crucial to industry productivity. In parallel, the new ‘digital biology’ era provides yet more and more data to find, analyse and exploit. In this chapter we look at the contribution that Semantic MediaWiki (SMW) technology has made to meeting the information challenges faced by Pfizer. We describe two use-cases that highlight the flexibility of this software and the ultimate benefit to the user.
17.1 The Targetpedia
Although many debate the relative merits of target-driven drug discovery [1], drug targets themselves remain a crucial pillar of pharmaceutical research. Thus, one of the most basic needs in any company is for a drug target reference, an encyclopaedia providing key facts concerning these important entities. In the majority of cases ‘target’ can of course, be either a single protein or some multimeric form such as a complex, protein-protein interaction or pharmacological grouping such as ‘calcium channels’ [2]. Consequently, the encyclopedia should include all proteins (whether or not they were established targets) and known multiprotein targets from humans and other key organisms. Although there are a number of sites on the internet that provide some form of summary information in this space (e.g. Wikipedia [3], Ensembl [4], GeneCards [5], EntrezGene [6]), none are particularly tuned to presenting the information most relevant for drug discovery users. Furthermore, such systems do not incorporate historical company-internal data, clearly something of high value when assessing discovery projects. Indeed, in large companies working on 50 + new targets per year, the ability to track the target portfolio and associated assets (milestones, endpoints, reagents, etc.) is vital. Targets are not unique to one disease area and access to compounds, clones, cell-lines and reagents from existing projects can rapidly accelerate new studies. More importantly, access to existing data on safety, chemical matter quality, pathways/mechanisms and biomarkers can ‘make or break’ a new idea. Although this is often difficult for any employee, it is particularly daunting for new colleagues lacking the IT system knowledge and people networks often required to find this material. Thus, the justification for a universal protein/target portal at Pfizer was substantial, forming an important component in an overall research information strategy.
17.1.1 Design choices
In 2010, Pfizer informaticians sought to design a replacement for legacy protein/target information systems within the company. Right from the start it was agreed that the next system should not merely be a cosmetic update, but address the future needs of the evolving research organisation. We therefore engaged in a series of interviews across the company, consulting colleagues with a diverse set of job functions, experience and grades. Rather than focusing on software tools, the interviews centred around user needs and asked questions such as:
![]() What data and information do you personally use in your target research?
What data and information do you personally use in your target research?
![]() Where do you obtain that data and information?
Where do you obtain that data and information?
![]() Where, if any place, do you store annotations or comments about the work you have performed on targets?
Where, if any place, do you store annotations or comments about the work you have performed on targets?
![]() Approximately how many targets per year does your Research Unit (RU) have in active research? How many are new?
Approximately how many targets per year does your Research Unit (RU) have in active research? How many are new?
![]() What are your pain points around target selection and validation?
What are your pain points around target selection and validation?
Based on these discussions, we developed a solid understanding of what the user community required from the new system and the benefits that these could bring. Highlights included:
![]() Use of internet tools: most colleagues described regular use of the internet as a primary mechanism to gain information on a potential target. Google and Wikipedia searches were most common, their speed and simplicity fitting well with a scientist’s busy schedule. The users knew there were other resources out there, but were unclear as to which were the best for a particular type of data, or how to stay on top of the ever-increasing number of relevant web sites.
Use of internet tools: most colleagues described regular use of the internet as a primary mechanism to gain information on a potential target. Google and Wikipedia searches were most common, their speed and simplicity fitting well with a scientist’s busy schedule. The users knew there were other resources out there, but were unclear as to which were the best for a particular type of data, or how to stay on top of the ever-increasing number of relevant web sites.
![]() Data diversity: many interviewees could describe the types of data that they would like to see in the system. Although there were some differences between scientists from different research areas (discussed later), many common areas emerged. Human genetics and genomic variation, disease association, model organism biology, expression patterns, availability of small molecules and the competitor landscape were all high on the list. Given that we identified an excess of 40 priority internal and external systems in these areas, there was clearly a need to help organise these data for Pfizer scientists.
Data diversity: many interviewees could describe the types of data that they would like to see in the system. Although there were some differences between scientists from different research areas (discussed later), many common areas emerged. Human genetics and genomic variation, disease association, model organism biology, expression patterns, availability of small molecules and the competitor landscape were all high on the list. Given that we identified an excess of 40 priority internal and external systems in these areas, there was clearly a need to help organise these data for Pfizer scientists.
![]() Accessing the internal target portfolio: many in the survey admitted frustration in accessing information on targets the company had previously studied. Such queries were possible, particularly via the corporate portfolio management platforms. However, these were originally designed for project management and other business concerns, and could not support more ‘molecular’ questions such as ‘Show me all the company targets in pathway X or gene family Y’. As expected, this issue was particularly acute for new colleagues who had not yet developed ‘work arounds’ to try to compile this type of information.
Accessing the internal target portfolio: many in the survey admitted frustration in accessing information on targets the company had previously studied. Such queries were possible, particularly via the corporate portfolio management platforms. However, these were originally designed for project management and other business concerns, and could not support more ‘molecular’ questions such as ‘Show me all the company targets in pathway X or gene family Y’. As expected, this issue was particularly acute for new colleagues who had not yet developed ‘work arounds’ to try to compile this type of information.
![]() Capturing proteins ‘of interest’: one of the most interesting findings was the desire to track proteins of interest to Pfizer scientists but that were not (yet) part of the official research portfolio. This included emerging targets that project teams were monitoring closely as well as proteins that were well established within a disease, but were not themselves deemed ‘druggable’. For instance, a transcription factor might regulate pathways crucial to two distinct therapeutic areas, yet as the teams are geographically distributed they may not realise a common interest. Indeed, many respondents cited this intersection between biological pathways and the ‘idea portfolio’ as an area in need of much more information support.
Capturing proteins ‘of interest’: one of the most interesting findings was the desire to track proteins of interest to Pfizer scientists but that were not (yet) part of the official research portfolio. This included emerging targets that project teams were monitoring closely as well as proteins that were well established within a disease, but were not themselves deemed ‘druggable’. For instance, a transcription factor might regulate pathways crucial to two distinct therapeutic areas, yet as the teams are geographically distributed they may not realise a common interest. Indeed, many respondents cited this intersection between biological pathways and the ‘idea portfolio’ as an area in need of much more information support.
![]() Embracing of ‘wikis’: we also learnt that a number of research units (RUs) had already experimented with their own approaches to managing day-to-day information on their targets and projects. Many had independently arrived at the same solution and implemented a wiki system. Users liked the simplicity, ease of collaboration and familiarity (given their use of Wikipedia and Pfizer’s own internal wiki, see Chapter 13 by Gardner and Revell). This highlighted that many colleagues were comfortable with this type of approach and encouraged us to develop a solution that was in tune with their thinking.
Embracing of ‘wikis’: we also learnt that a number of research units (RUs) had already experimented with their own approaches to managing day-to-day information on their targets and projects. Many had independently arrived at the same solution and implemented a wiki system. Users liked the simplicity, ease of collaboration and familiarity (given their use of Wikipedia and Pfizer’s own internal wiki, see Chapter 13 by Gardner and Revell). This highlighted that many colleagues were comfortable with this type of approach and encouraged us to develop a solution that was in tune with their thinking.
![]() Annotation: responses concerning annotation were mixed; there were a range of opinions as to whether the time spent adding comments and links to information pages was a valuable use of a scientists’ time. We also found that users viewed existing annotation software as too cumbersome, often requiring many steps and logging in and out of multiple systems. However, it was clear from the RU-wikis that there was some appetite for community editing when placed in the right context. It became very clear that if we wanted to encourage this activity we needed to provide good tools that were a fit with the scientists’ workflow and provided a tangible value to them.
Annotation: responses concerning annotation were mixed; there were a range of opinions as to whether the time spent adding comments and links to information pages was a valuable use of a scientists’ time. We also found that users viewed existing annotation software as too cumbersome, often requiring many steps and logging in and out of multiple systems. However, it was clear from the RU-wikis that there was some appetite for community editing when placed in the right context. It became very clear that if we wanted to encourage this activity we needed to provide good tools that were a fit with the scientists’ workflow and provided a tangible value to them.
17.1.2 Building the system
The Targetpedia was intended to be a ‘first stop’ for target-orientated information within the company. As such, it needed to present an integrated view across a range of internal and external data sources. The business analysis provided a clear steer towards a wiki-based system and we reviewed many of the different platforms before deciding on the Semantic MediaWiki (SMW) framework [7]. SMW (which is an extension of the MediaWiki software behind Wikipedia) was chosen for the following reasons:
![]() Agility: although our interviews suggested users would approve of the wiki approach, we were keen to produce a prototype to test this hypothesis. SMW offered many of the capabilities we needed ‘out of the box’ – certainly enough to produce a working prototype. In addition, the knowledge that this same software underlies both Wikipedia and our internal corporate wiki suggested that (should we be successful), developing a production system should be possible.
Agility: although our interviews suggested users would approve of the wiki approach, we were keen to produce a prototype to test this hypothesis. SMW offered many of the capabilities we needed ‘out of the box’ – certainly enough to produce a working prototype. In addition, the knowledge that this same software underlies both Wikipedia and our internal corporate wiki suggested that (should we be successful), developing a production system should be possible.
![]() Familiarity: as the majority of scientists within the company were familiar with MediaWiki-based sites, and many of our specific target customers had set up their own instances, we should not face too high a barrier for adopting a new system.
Familiarity: as the majority of scientists within the company were familiar with MediaWiki-based sites, and many of our specific target customers had set up their own instances, we should not face too high a barrier for adopting a new system.
![]() Extensibility: although SMW had enough functionality to meet early stage requirements, we anticipated that eventually we would need to extend the system. The open codebase and modular design were highly attractive here, allowing our developers to build new components as required and enabling us to respond to our customers quickly.
Extensibility: although SMW had enough functionality to meet early stage requirements, we anticipated that eventually we would need to extend the system. The open codebase and modular design were highly attractive here, allowing our developers to build new components as required and enabling us to respond to our customers quickly.
![]() Semantic capabilities: a key element of functionality was the ability to provide summarisation and taxonomy-based views across the proteins (described in detail below). This is actually one of the most powerful core capabilities of SMW and something not supported by many of the alternatives. The feature is enabled by the ‘ASK’ query language [8], which functions somewhat like SQL and can be embedded within wiki pages to create dynamic and interactive result sets.
Semantic capabilities: a key element of functionality was the ability to provide summarisation and taxonomy-based views across the proteins (described in detail below). This is actually one of the most powerful core capabilities of SMW and something not supported by many of the alternatives. The feature is enabled by the ‘ASK’ query language [8], which functions somewhat like SQL and can be embedded within wiki pages to create dynamic and interactive result sets.
Data sourcing
Using a combination of user guidance and access statistics from legacy systems, we identified the major content elements required for the wiki. For version one of Targetpedia, the entities chosen were: proteins and protein targets, species, indications, pathways, biological function annotations, Pfizer people, departments, projects and research units.
For each entity we then identified the types and sources of data the system needed to hold. Table 17.1 provides an excerpt of this analysis for the protein/target entity type. In particular, we made use of our existing infrastructure for text-mining of the biomedical literature, Pharmamatrix (PMx, [9]). PMx works by automated, massive-scale analysis of Medline and other text sources to identify associations between thousands of biomedical entities. The results of this mining provide a rich data source to augment many of the areas of scientific interest.
Table 17.1
Protein information sources for Targetpedia
| Name | Description | Example sources |
| General functional information | General textual summary of the target | Wikipedia [3] EntrezGene [6] Gene Ontology [10] |
| Chromosomal information | Key facts on parent gene within genome | Ensembl [4] |
| Disease links | Known disease associations | Genetic Association Database [11] OMIM [12] Literature associations (PMx) GWAS data (e.g. [13]) |
| Genetics | Polymorphisms and mouse phenotypes | Polyphen 2 [14] Mouse Genome Informatics [15] Internal data Literature mining (PMx) |
| Drug discovery | Competitor landscape for this target | Multiple commercial competitor databases [2] |
| Safety | Safety issues | Comparative Toxicogenomics Database [16] Pfizer safety review repository Literature mining (PMx) |
| Pfizer project information | Projects have we worked on this target for | Discovery and Development portfolio databases Pfizer screening registration database |
| References | Key references | Gene RIFs [6] Literature mining (PMx) |
| Pathways | Pathways is this protein a member of | Reactome [17] BioCarta [18] Commercial pathway systems |
Loading data
Having identified the necessary data, a methodology for loading this into the Targetpedia was designed and implemented, schematically represented in Figure 17.1 and discussed below.
As Chapter 16 by Alquier describes the core SMW system in detail, here we will highlight the important Targetpedia-specific elements of the architecture, such as:
![]() Source management: for external sources (public and licensed commercial) a variety of data replication and scheduling tools were used (including FTP, AutoSys [19] and Oracle materialised views) to manage regular updates from source into our data warehouse. Most data sets are then indexed by and made queryable by loading into Oracle, Lucene [20] or SRS [21]. Data sources use a vast array of different identifiers for biomedical concepts such as genes, proteins and diseases. We used an internal system (similar to systems such as BridgeDb [22]) to provide mappings between different identifiers for the same entity. Multiprotein targets were sourced from our previously described internal drug target database [2]. Diseases were mapped to our internal disease dictionary, which is an augmented form of the disease and condition branches of MeSH [23].
Source management: for external sources (public and licensed commercial) a variety of data replication and scheduling tools were used (including FTP, AutoSys [19] and Oracle materialised views) to manage regular updates from source into our data warehouse. Most data sets are then indexed by and made queryable by loading into Oracle, Lucene [20] or SRS [21]. Data sources use a vast array of different identifiers for biomedical concepts such as genes, proteins and diseases. We used an internal system (similar to systems such as BridgeDb [22]) to provide mappings between different identifiers for the same entity. Multiprotein targets were sourced from our previously described internal drug target database [2]. Diseases were mapped to our internal disease dictionary, which is an augmented form of the disease and condition branches of MeSH [23].
![]() Data provision: for each source, queries required to obtain information for the wiki were identified. In many instances this took the form of summaries and aggregations rather than simply extracting data ‘as-is’ (described below). Each query was turned into a REST-ful web service via an in-house framework known as BioServices. This acts a wrapper around queries and takes care of many common functions such as load balancing, authentication, output formatting and metadata descriptions. Consequently, each query service (‘QWS’ in the figure) is a standardised, parameterised endpoint that provides results in a variety of formats including XML and JSON. A further advantage of this approach is that these services (such as ‘general information for a protein’, ‘ontology mappings for a protein’) are also available outside of Targetpedia.
Data provision: for each source, queries required to obtain information for the wiki were identified. In many instances this took the form of summaries and aggregations rather than simply extracting data ‘as-is’ (described below). Each query was turned into a REST-ful web service via an in-house framework known as BioServices. This acts a wrapper around queries and takes care of many common functions such as load balancing, authentication, output formatting and metadata descriptions. Consequently, each query service (‘QWS’ in the figure) is a standardised, parameterised endpoint that provides results in a variety of formats including XML and JSON. A further advantage of this approach is that these services (such as ‘general information for a protein’, ‘ontology mappings for a protein’) are also available outside of Targetpedia.
![]() Data loading: a MediaWiki ‘bot [24] was developed to carry out the loading of data into Targetpedia. A ‘bot is simply a piece of software that can interact with the MediaWiki system to load large amounts of content directly into the backend database. Each query service was registered with the ‘bot, along with a corresponding wiki template that would hold the actual data. Templates [25] are similar to classes within object-orientated programming in that they define the fields (properties) of a complex piece of data. Critically, templates also contain a definition of how the data should be rendered in HTML (within the wiki). This approach separates the data from the presentation, allowing a flexible interface design and multiple views across the same object. With these two critical pieces in place, the ‘bot was then able to access the data and load it into the system via the MediaWiki API [26].
Data loading: a MediaWiki ‘bot [24] was developed to carry out the loading of data into Targetpedia. A ‘bot is simply a piece of software that can interact with the MediaWiki system to load large amounts of content directly into the backend database. Each query service was registered with the ‘bot, along with a corresponding wiki template that would hold the actual data. Templates [25] are similar to classes within object-orientated programming in that they define the fields (properties) of a complex piece of data. Critically, templates also contain a definition of how the data should be rendered in HTML (within the wiki). This approach separates the data from the presentation, allowing a flexible interface design and multiple views across the same object. With these two critical pieces in place, the ‘bot was then able to access the data and load it into the system via the MediaWiki API [26].
![]() Update scheduling: as different sources update content at different rates, the system was designed such that administrators could refresh different elements of the wiki under different schedules. Furthermore, as one might often identify errors or omissions affecting only a subset of entries, the loader was designed such that any update could be limited to a list of specific proteins if desired. Thus, administrators have complete control over which sources and entries can be updated at any particular time.
Update scheduling: as different sources update content at different rates, the system was designed such that administrators could refresh different elements of the wiki under different schedules. Furthermore, as one might often identify errors or omissions affecting only a subset of entries, the loader was designed such that any update could be limited to a list of specific proteins if desired. Thus, administrators have complete control over which sources and entries can be updated at any particular time.
The result of this process was a wiki populated with data covering the major entity types in a semantically enriched format. Figure 17.2 shows an excerpt of the types of properties and values stored for the protein, phosphodiesterase 5.
17.1.3 The product and user experience
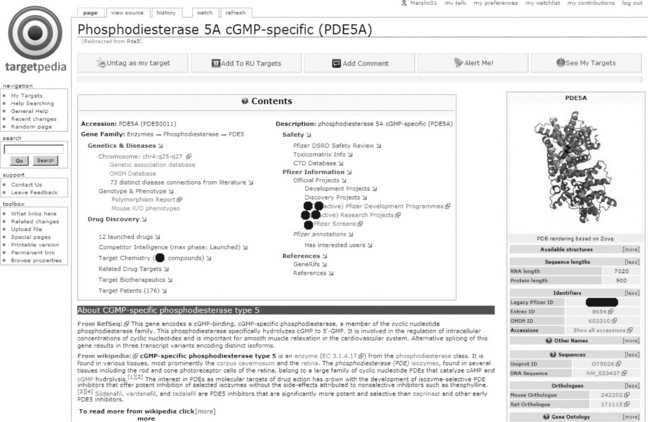
The starting point for most users is the protein information pages. Figure 17.3 shows one such example.
The protein pages have a number of important design features.
![]() Social tagging: at the top of every page, buttons that allow users to ‘tag’ and annotate targets of interest (discussed in detail below).
Social tagging: at the top of every page, buttons that allow users to ‘tag’ and annotate targets of interest (discussed in detail below).
![]() The InfoBox: as commonly used within Wikipedia pages, customised to display key facts, including database identifiers, synonyms, pathways and Gene Ontology processes in which the protein participates.
The InfoBox: as commonly used within Wikipedia pages, customised to display key facts, including database identifiers, synonyms, pathways and Gene Ontology processes in which the protein participates.
![]() Informative table of contents: the default MediaWiki table of contents was replaced with a new component that combines an immediate, high-level summary of critical information with hyperlinks to relevant sections.
Informative table of contents: the default MediaWiki table of contents was replaced with a new component that combines an immediate, high-level summary of critical information with hyperlinks to relevant sections.
![]() Textual overview: short, easily digestible summaries around the role and function of this entity are provided. Some of this content is obtained directly from Wikipedia, an excellent source thanks to the Gene Wiki initiative [27]. For this, the system only presents the first one or two paragraphs initially; the section can be expanded to the full article with a single click. This is complemented with textual summaries from the National Center for Biotechnology Information’s RefSeq [28] database.
Textual overview: short, easily digestible summaries around the role and function of this entity are provided. Some of this content is obtained directly from Wikipedia, an excellent source thanks to the Gene Wiki initiative [27]. For this, the system only presents the first one or two paragraphs initially; the section can be expanded to the full article with a single click. This is complemented with textual summaries from the National Center for Biotechnology Information’s RefSeq [28] database.
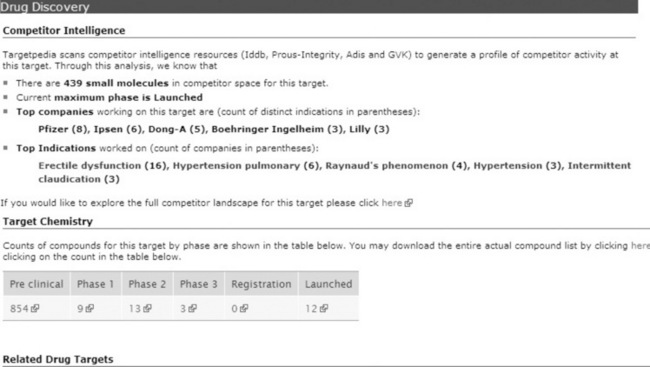
The rest of the page is made up of more detailed sections covering the areas described in Table 17.1. For example, Figure 17.4 shows the information we display in the competitor intelligence section and demonstrates how a level of summarisation has been applied. Rather than presenting all of the competitor data, we present some key facts such as number of known small molecules, maximum clinical phase and most common indications. Should the user be sufficiently interested to know more, many of the elements in the section are hyperlinked to a more extensive target intelligence system [2], which allows them to analyse all of the underlying competitor data.
One of the most powerful features of SMW is the ability to ‘slice and dice’ content based on semantic properties, allowing developers (and even technically savvy users) to create additional views of the information. For example, each protein in the system has a semantic property that represents its position within a global protein family taxonomy (based on a medicinal chemistry view of protein function, see [29]). Figure 17.5 shows how this property can be exploited though an ASK query to produce a view of proteins according to their family membership, in this case the phosphodiesterase 4 family. The confidential information has been blocked out, Pfizer users see a ‘dashboard’ for the entire set of proteins and to what extent they have been investigated by the company. Critically, SMW understands hierarchies, so if the user were to view the same page for ‘phosphodiesterases’ they would see all such proteins, including the four shown here. Similar views have been set up for pathway, disease and Gene Ontology functions, providing a powerful mechanism for looking across the data – and all (almost), for free!
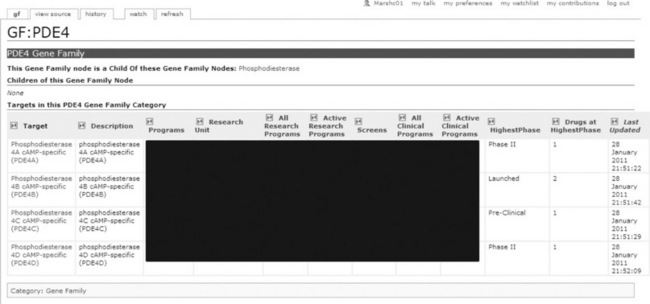
Figure 17.5 A protein family view. Internal users would see the table filled with company data regarding progression of each protein within drug discovery programmes
Collaboration in Targetpedia
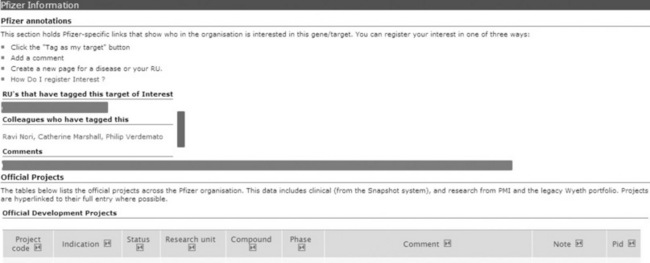
One of the major differences between Targetpedia and our legacy protein/ target information systems are features that empower internal collaboration. For instance, Pfizer drug discovery project tracking codes are found on all pages that represent internal targets, providing an easy link to business data. Each project code has its own page within Targetpedia, listing the current status, milestones achieved and, importantly, the people associated with the research. The connection of projects to scientists was made possible thanks to the corporate timesheets that all Pfizer scientists complete each week, allocating their time against specific project codes. By integrating this into Targetpedia, this administrative activity moves from simply a management tool to something that tangibly enables collaboration. Further integration with departmental information systems provides lists of colleagues involved in the work, organised by work area. This helps users of Targetpedia find not just the people involved, but those from say, the pharmacodynamics or high-throughput screening groups. We believe this provides a major advance in helping Pfizer colleagues find the right person to speak to regarding a project on a target in which they have become interested.
A second collaboration mechanism revolves around the concept of the ‘idea portfolio’. We wanted to make it very easy for users to assert an interest in a particular protein. Similar to the Facebook ‘like’ function, the first button in the tagging bar (Figure 17.3) allows users to ‘Tag (this protein) as my target’ with a single click. This makes it trivial for scientists to create a portfolio of proteins of interest to them or their research unit. An immediate benefit is access to a range of alerting tools, providing email or RSS updates to new updates, database entries or literature regarding their chosen proteins. However, the action of tagging targets creates a very rich data set that can be exploited to identify connections between disparate individuals in a global organisation. Interest in the same target is obvious, but as mentioned above, algorithms that scan the connections to identify colleagues with interests in different proteins but the same pathway present very powerful demonstrations of the value of this social tagging.
Finally, Targetpedia is of course, a wiki, and whereas much of the above has concerned the import, organisation and presentation of existing information, we wanted to also enable the capture of pertinent content from our users. This is, in fact, more complex than it might initially seem. As described in the business analysis section, many research units were running their own wikis, annotating protein pages with specific project information. However, this is somewhat at odds with a company-wide system, where different units working on the same target would not want their information to be merged. Furthermore, pages containing large sections of content that are specifically written by or for a very limited group of people erodes the ‘encyclopaedia’ vision for Targetpedia. To address this, it was decided that the main protein pages would not be editable, allowing us to retain the uniform structure, layout and content across all entries. This also meant that updating would be significantly simpler, not having to distinguish between user-generated and automatically loaded content.
To provide wiki functionality, users can (with a single click) create new ‘child’ pages that are automatically and very clearly linked to the main page for that protein. These can be assigned with different scopes, such as a user-page, for example ‘Lee Harland’s PDE5 page’, a project page, for example ‘The Kinase Group Page For MAPK2’ and a research unit page, for example ‘Oncology p53 programme page’. In this way, both the encyclopaedia and the RU-specific information capture aspects of Targetpedia are possible in the same system. Search results for a protein will always take a user to a main page, but all child pages are clearly visible and accessible from this entry point. At present, the template for the subpages is quite open, allowing teams to build those pages as they see fit, in keeping with their work in their own wiki systems. Finally, there may be instances where people wish to add very short annotations – a key paper or a URL that points to some useful information. For this, the child page mechanism may be overkill. Therefore, we added a fourth collaborative function that allows users to enter a short (255 character) message by clicking the ‘add comment’ button on the social tagging toolbar. Crucially, the messages are not written into the page itself, but stored within the SMW system and dynamically included via an embedded ASK query. This retains the simple update pattern for the main page and allows for modification to the presentation of the comments in future versions.
17.1.4 Lessons learned
Targetpedia has been in use for over six months, with very positive feedback. During this time there has been much organisational change for Pfizer, re-affirming the need for a central repository of target information. Yet, in comparison with our legacy systems, Targetpedia has changed direction in two major areas. Specifically, it shifts from a ‘give me everything in one go’ philosophy to ‘give me a summary and pointers where to go next’. Additionally, it addresses the increasing need for social networking, particularly through shared scientific interest in the molecular targets themselves.
Developing with SMW was generally a positive experience, so much so we went on to re-use components in a second project described below. Templating in particular is very powerful, as are the semantic capabilities that make this system unique within its domain. Performance (in terms of rendering the pages) was never an issue, although we did take great care to optimise the semantic ASK queries by limiting the number of joins across different objects in the wiki. However, the speed by which content could be imported into the system was something that was suboptimal. There is a considerable amount of data for over 20 000 proteins, as well as people, diseases, projects and other entities. Loading all of this via the MediaWiki API took around a day to perform a complete refresh. As the API performs a number of operations in addition to loading the MySQL database, we could not simply bypass it and insert content directly into the database itself. Therefore, alternatives to the current loading system may have to be found for the longer term, something that will involve detailed analysis of the current API. Yet, even with this issue, we were able to provide updates along a very good timeline that was acceptable to the user community. A second area of difficulty was correctly configuring the system for text searches. The MediaWiki search engine is quite particular in how it searches the system and displays the results, which forced us to alter the names of many pages so search results returned titles meaningful to the user.
Vocabulary issues represented another major hurdle, and although not the ‘fault’ of SMW, they did hinder the project. For instance, there are a variety of sources that map targets to indications (OMIM, internal project data, competitor intelligence), yet each uses a different disease dictionary. Thus, users wish to click on ‘asthma’ to see all associated proteins and targets, but this is quite difficult to achieve without much laborious mapping of disease identifiers and terms. Conversely, there are no publically available standards around multi-protein drug targets (e.g. ‘protein kinase C’, ‘GABA receptor’, ‘gastric proton pump’). This means that public data regarding these entities are poorly organised and difficult to identify and integrate. As a consequence, we are able to present only a small amount of information for these entities, mostly driven from internal systems (we do, of course, provide links to the pages for the individual protein components). We believe target information provision would be much enhanced if there were open standards and identifiers available for these entities. Indeed, addressing vocabulary and identity challenges may be key to progressing information integration and appear to be good candidates for pre-competitive collaboration [30].
For the future, we would like to create research unit-specific sections within the pages, sourced from (e.g.) disease-specific databases and configured to appear to relevant users only. We are also investigating the integration of ‘live’ data by consuming web services directly into the wiki using available extensions [31]. We conclude that SMW provides a powerful platform from which to deliver this and other enhancements for our users.
17.2 The Disease Knowledge Workbench (DKWB)
The Pfizer Indications Discovery Research Unit (IDU) was established to identify and explore alternative indications for compounds that have reached late stage clinical development. The IDU is a highly collaborative group; project leads, academic collaborators and computational scientists work together to develop holistic views of the cellular processes and molecular pathways within a disease of interest. Such deep understanding allows the unit to address key areas such as patient stratification, confidence in target rationale and identification of the best therapeutic mechanism and outcome biomarkers. Thus, fulfilling the IDUs’ mission requires the effective management and exploitation of relevant information across a very diverse range of diseases.
The information assessed by the IDU comes from a range of internal and external sources, covering both raw experimental output and higher-level information such as the biomedical literature. Interestingly, the major informatics gap was not the management and mining of these data per se, as these functions were already well provided for with internal and public databases and tools. Rather, the group needed a mechanism to bring together and disseminate the knowledge that they had assessed and synthesised, essentially detailing their hypotheses and the data that led them there. As one would expect, the scientists used lab note books to record experiments, data mining tools to analyse data and read many scientific papers. However, like many organisations, the only place in which each ‘story’ was brought together was in a slide deck, driven by the need to present coherent arguments to the group members and management. Of course, this is not optimal with these files (including revisions and variants) quickly becoming scattered through hard drives and document management systems. Furthermore, they often lack clear links to data supporting conclusions and have no capability to link shared biology across different projects. This latter point can be especially critical in a group running a number of concurrent investigations; staying abreast of the major elements of each project and their interdependencies can be difficult. Therefore, the IDU and the computational sciences group began an experiment to develop tools to move disease knowledge management away from slides and into a more fit for purpose environment.
17.2.1 Design choices
Any piece of software that allows users to enter content could fall into the category of ‘knowledge management’. However, there are a number of tools that allow users to represent coherent stories in an electronic representation. These range from notebook-style applications [32] to general mind-mapping software (e.g. [33]) and more specialised variants such as the Compendium platform for idea management [34]. A particularly relevant example is the I2 Analysts Notebook [35], an application used throughout law enforcement, intelligence and insurance agencies to represent complex stories in semi-graphical form. Although we had previously explored this for knowledge representation [2] and were impressed with its usability and relationship management, its lack of ‘tuning’ to the biomedical domain was a major limiting factor.
A further type of system considered was one very tuned to disease modelling, such as the Biological Expression Language (BEL) Framework produced by Selventa [36]. This approach describes individual molecular entities in a causal network, allowing computer modelling and simulation to be performed to understand the effects of any form of perturbation. Although this is an important technology, it works at a very different level to the needs of the IDU. Indeed, our aim was not to try to create a mathematical or causal model of the disease, but to provide somewhere for the scientists interpretation of these and other evidence to reside. This interpretation takes the form of a ‘report’ comprising free text, figures/images, tables, text and hyperlinks to files of underlying evidence. This is, of course, very wiki-like, and the system which most matched these needs was AlzSWAN [37], a community-driven knowledge base of Alzheimer disease hypotheses. AlzSWAN is a wiki-based platform that allows researchers to propose, annotate, support and refute particular pathological mechanisms. Users attach ‘statements’ to a hypothesis (based on literature, experimental data or their own ideas), marking whether these are consistent with the hypothesis. This mechanism to present, organise and discuss the pathophysiological elements of the disease was very aligned with the needs of our project, capturing at the level of scientific discourse, rather than individual molecular networks.
17.2.2 Building the system
Although AlzSWAN came closest to our desired system, it has a top-down approach to knowledge management; one provides a hypothesis and then provides the evidence for and against it. In the case of the IDU, the opposite was required, namely the ability to manage individual findings and ideas with the ultimate aim of bringing these together within a final therapeutic strategy. Thus, we concluded that a knowledge structure different from AlzSWAN was needed, but still took great inspiration from that tool. Given that the concept of the disease knowledge workbench was very much exploratory, a way to develop a prototype at minimal cost was essential. It was here that experience with SMW within Targetpedia enabled the group to move rapidly and create an AlzSWAN-like system, tailored to the IDU’s needs, within a matter of weeks. Without the ready availability of a technology that could provide the basis for the system, it is unlikely that the idea of building the workbench would have turned into a reality.
Modelling information in DKWB
Informaticians and IDU scientists collaborated in developing the information model required to correctly manage disease information in the workbench. The principle need was to take a condition such as sepsis and divide it into individual physiological components, as shown in Figure 17.7.
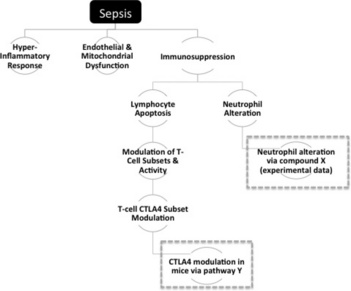
Figure 17.7 Dividing sepsis into physiological subcomponents. Dashed grey boxes surround components that are hypothesised and not yet validated by the team
Each component in Figure 17.7 represents an ‘assertion’, something that describes a specific piece of biology within the overall context of the disease. As can be seen, these are not simple binary statements, but represent more complex scientific phenomena, an example being ‘increases in levels of circulating T-Lymphocytes are primarily the result of the inflammatory cascade’. There are multiple levels of resolution and assertions can be parents or children of other assertions. For instance, assertions can be observations (biology with supporting experimental or published data) or hypotheses (predicted but not yet experimentally supported). It should be noted that just because observations were the product of published experiments that does not necessarily mean that they are treated as fact. Indeed, one of the primary uses for the system was to critique published experiments and store the teams view of whether that data could be trusted. Regardless, each can be associated with a set of properties covering its nature and role within the system, as described in Table 17.2. Properties such as the evidence level are designed to allow filtering between clinical and known disease biology and more unreliable data from pre-clinical studies.
Table 17.2
The composition of an assertion
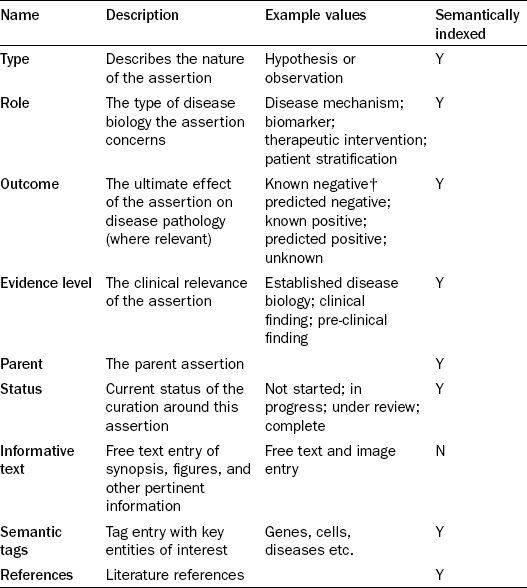
†in this context, ‘negative’ means making the disease or symptoms worse
Although we did not want to force scientists to encode extensive complex interpretations into a structured form, there was a need to identify and capture the important components of any assertion. Thus, a second class of concepts within the workbench were created, known as ‘objects’. An object represents a single biomedical entity such as a gene, protein, target, drug, disease, cellular process and so on. Software developed on the Targetpedia project allowed us to load sets of known entities from source databases and vocabularies into the system, in bulk. By themselves, these objects are ‘inert’, but they come alive when semantically tied to assertions. For instance, for the hypothesis ‘Altered monocyte cytokine release contributes to immunosuppression in sepsis’, one can attach the entities involved such as monocyte cells and specific cytokine proteins. This ‘semantic tagging’ builds up comprehensive relationships between pathophysiology and the cellular and molecular components that underlie these effects. They also enable the identification of shared biology. For instance, the wiki can store all known molecular pathways as observations; assigning all of the protein members to it via semantic tagging. Then, as authors of other assertions tag their pages with the principle proteins involved, an ASK query can automatically identify relevant pathways and present them on the page without any need for user intervention. This can be extended to show all assertions in the system that are semantically tagged with proteins in any shared pathway, regardless of what disease areas they describe. This aids the identification of common pathways and potential synergies across projects.
17.2.3 The product and user experience
To allow Pfizer scientists to fully describe and present their hypotheses and summaries, the assertions within the system are a mix of structured data (Table 17.2, semantic tagging) and free-form text and images. The semantic forms extension [38] provided a powerful and elegant mechanism to structure this data entry. Figure 17.8 shows how an assertion is created or edited.

Figure 17.8 The Semantic Form for creating a new assertion. Note, the free-text entry area (under ‘Main Document Text’) has been truncated for the figure
The top of the edit form contains a number of drop-down menus (dynamically built from vocabularies in the system) to enable users to rapidly set the main properties for an assertion. Below this is the main wiki-text editing area, using the FCKEditor plug-in that enables users to work in a word-processor like mode without needing to learn the wiki markup language. Next is the semantic tagging area, where assertions can be associated with the objects of interest. Here, the excellent auto-complete mechanism that suggests as one types is employed, allowing the user to more easily enter the correct terms from supported vocabularies. The user must enter the nature of the association (‘Link’ on the figure), chosen from a drop-down menu. One of the most common links is ‘Has Actor’, meaning that the assertion is associated with a molecular entity that plays an active role within it. Finally, additional figures and references can be added using the controls at the bottom of the page.
Once the user has finished editing, the page is rendered into a user-friendly viewing format via a wiki template, as shown in Figure 17.9(a). As can be seen in Figure 17.9(b), the system renders semantic tags (in this case showing that CD4 + T-cells have been associated with the assertion). In addition, via an ASK query, connections to other assertions in the system are identified and dynamically inserted at the bottom of the page, in this case identifying two child observations that already exist in the system.

Figure 17.9 (a) An assertion page as seen after editing. (b) A semantic tag and automatic identification of related assertions
All assertions are created as part of a ‘project’, normally focused on a disease of interest. In some cases disease knowledge was retrospectively entered into the system from legacy documents and notes and subsequently further expanded. New projects were initiated directly in the wiki, following discussions on the major elements of the disease by the scientist.
From the main project page (Figure 17.10), one can get an overview of the disease, its pathogenesis and progression, diagnosis and prevalence, standard care and market potential, as well as key disease mechanisms. The assertions listed at the bottom of this page (Figure 17.10(b) ) are divided into three sections: (1) ‘top level disease mechanism’ (i.e. those directly linked to the page); (2) ‘all other disease mechanisms below the top level’ (i.e. those which are children of parent associations in (1)); and (3) ‘non-disease mechanism assertions such as target/pathway or therapeutic intervention’. One can drill into one of the assertion pages and from there navigate back to the project main page or any parent or child assertion pages using the semantic network hyperlinks. In this way the workbench is not only useful as a way to capture disease knowledge but also provides a platform to present learning on diseases and pathways to project teams across the company. As individual targets and diseases are semantically indexed on the pages, one can explore the knowledge base in many ways and move freely from one project to another.

Figure 17.10 The sepsis project page. The upper portion (a) is generated by the user. The lower portion (b) is created automatically by the system
In summary, we have developed a knowledge system that allows scientists to collaboratively build and share disease knowledge and interpretation with a low barrier to adoption. Much of the data capture process is deliberately manual – aiming to capture what the scientist thinks, rather than what a database knows. However, future iterations of the tool could benefit from more intelligent and efficient forms of data entry, annotation and discovery. Better connectivity to internal databases would be a key next step – as assertions are entered, they could be scanned and connected to important results in corporate systems. Automatic semantic tagging would be another area for improvement, the system recognising entities as the text is typed and presenting the list of ‘discovered’ concepts to the user who simply checks it is correct. Finally, we envisage a connection with tools such as Utopia (see Chapter 15 by Pettifer and colleagues), that would allow scientists to augment their assertions by sending quotes and comments on scientific papers directly from the PDF reader to the wiki.
17.2.4 Lessons learned
The prototype disease knowledge workbench brings a new capability to our biologists, enabling the capture of their understanding and interpretation of disease mechanisms. However, there are definitely areas for improvement. In particular, although the solution was based around the writing of report-style content, the MediaWiki software lacks a robust, intuitive interface for general editing. The FCKEditor component provides some basic functionality, but it still feels as if it is not fully integrated and lacks many common features. Incorporation of images and other files needs to evolve to a ‘drag and drop’ method, rather than the convoluted, multistep process used currently. Although we are aware of the Semantic MediaWiki Plus extensions in these areas [39], in our hands they were not intuitive to use and added too much complexity to the interface. Thus, the development of components to make editing content much more user-friendly should be a high priority and one that, thanks to the open source nature, could be undertaken by ourselves and donated back to the community.
As for the general concept, the disease knowledge capture has many benefits and aids organisations in capture, retaining and (re-)finding the information surrounding key project decisions. However, a key question is, will scientists invest the necessary time to enter their knowledge into the system? Our belief is that scientists are willing to do this, when they see tangible value from such approaches. This requires systems to be built that combine highly user-orientated interfaces with intelligent semantics. Although we are not there yet, software such as SMW provides a tantalising glimpse into what could be in the future. Yet, as knowledge management technologies develop there is still a need to look at the cultural aspects of organisations and better mechanisms to promote and reward these valuable activities. Otherwise, organisations like Pfizer will be doomed to a life of representing knowledge in slides and documents, lacking the provenance required to trace critical statements and decisions.
17.3 Conclusion
We have deployed Semantic MediaWiki for two contrasting use-cases. Targetpedia is a system that combines large-scale data integration with social networking while the Disease Knowledge Workbench seeks to capture discourse and interpretation into a looser form. In both cases, SMW allowed us to accelerate the delivery of prototypes to test these new mechanisms for information management and converting tacit knowledge into explicit, searchable facts. This is undeniably important – as research moves into a new era where ‘data is king’, companies must identify new and better ways of managing those data. Yet, developers may not always know the requirements up front, and initial systems always require tuning once they are released into real-world use. Software that enables informatics to work with the science and not against it is crucial, and something that SMW arguably demonstrates perfectly. Furthermore, as described by Alquier (Chapter 16), SMW is geared to the semantic web, undoubtedly a major component of future information and knowledge management. SMW is not perfect, particularly in the area of GUI interaction, and like any open source technology some of the extensions were not well documented or had incompatibilities. However, it stands out as a very impressive piece of software and one that has significantly enhanced our information environment.
17.4 Acknowledgements
We thank members of the IDU, eBiology, Computational Sciences CoE and Research informatics for valuable contributions and feedback. In particular, Christoph brockel, Enoch Huang, Cory Brouwer, Ravi Nori, Bryn-Williams-Jones, Eric Fauman, Phoebe Roberts, Robert Hernandez, Markella Skempri. We would like to specifically acknowledge Michael Berry and Milena Skwierawska who developed much of the original Targetpedia codebase. We thank David Burrows & Nigel Wilkinson for data management and assistance in preparing the manuscript.
17.5 References
[1] Swinney, D., How were new medicines discovered? Nature Reviews Drug Discovery, 2011.
[2] Harland, L., Gaulton, A. Drug target central. Expert Opinion on Drug Discovery. 2009; 4:857–872.
[3] Wikipedia, http://en.wikipedia.org.
[4] Ensembl, http://www.ensembl.org.
[5] GeneCards, http://www.genecards.org/.
[6] EntrezGene, http://www.ncbi.nlm.nih.gov/gene.
[7] Semantic MediaWiki, http://www.semantic-mediawiki.org.
[8] Semantic MediaWiki Inline-Queries, http://semantic-mediawiki.org/wiki/Help:Inline_queries.
[9] Hopkins et al. System And Method For The Computer-Assisted Identification Of Drugs And Indications. US 2005/0060305.
[10] Harris, M.A., et al. Gene Ontology Consortium: The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004; 32:D258–61.
[11] Genetic Association Database, http://geneticassociationdb.nih.gov/.
[12] Online Mendelian Inheritance In Man (OMIM), http://www.ncbi.nlm.nih.gov/omim.
[13] Collins, F. Reengineering Translational Science: The Time Is Right. Science Translational Medicine. 2011.
[14] PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/.
[15] Mouse Genome Informatics, http://www.informatics.jax.org/.
[16] Comparative Toxicogenomics Database, http://ctd.mdibl.org/.
[17] Reactome, http://www.reactome.org/.
[18] Biocarta, http://www.biocarta.com/genes/index.asp.
[19] AutoSyshttp://www.ca.com/Files/ProductBriefs/ca-autosys-workld-autom-r11_p-b_fr_200711.pdf.
[20] Apache Lucene, http://lucene.apache.org/java/docs/index.html.
[21] Biowisdom SRS, http://www.biowisdom.com/2009/12/srs/.
[22] van Iersel, M.P., et al. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC Bioinformatics. 2010; 11:5.
[23] Medical Subject Headings (MeSH), http://www.nlm.nih.gov/mesh/.
[24] MediaWiki ‘Bot, http://www.mediawiki.org/wiki/HelpBots.
[25] MediaWiki Templates, http://www.mediawiki.org/wiki/HelpTemplates.
[26] MediaWiki API, http://www.mediawiki.org/wiki/API-.Main_page.
[27] Huss, J.W., et al. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Research. 2010; 38:D633–9.
[28] RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq/.
[29] Schuffenhauer, A., et al. An ontology for pharmaceutical ligands and its application for in silico screening and library design ). Journal of Chemical Information and Computer Sciences. 2002; 42:947–955.
[30] Harland, L., et al, Empowering Industrial Research with Shared Biomedical Vocabularies. Drug Discovery Today 2011;, doi: 10.1016/j. drudis.2011.09.013.
[31] MediaWiki Data Import Extension, http://www.mediawiki.org/wiki/Extension:Data_lmport_Extension.
[32] Barber, C.G., et al. ‘OnePoint–’ combining OneNote and SharePoint to facilitate knowledge transfer. Drug Discovery Today. 2009; 14:845–850.
[33] FreeMind, http://freemind.sourceforge.net/wiki/index.php/Main_Page.
[34] Compendium, http://compendium.open.ac.uk/institute/download/download.htm.
[35] i2 Analysts Notebook, http://www.i2group.com/us/products–-services/analysis-product-line/analysts-notebook.
[36] Selventa BEL Frameworkhttp://www.selventa.com/technology/bel-framework.
[37] AlzSwan, http://www.alzforum.org/res/adh/swan/default.asp.
[38] Semantic Forms, http://www.mediawiki.org/wiki/ExtensionSemantic_Forms.
[39] Semantic MediaWiki +, http://smwforum.ontoprise.com/smwforum.