Chapter 2. Case Study: Data Cleaning
Now that we know how to start RStudio, let’s dive in. We’ll begin with a blow-by-blow account of a sample data analysis for which we read in some data, clean it up, then format it for further study. We deliberately chose an example that will take us on some detours, as the point of the exercise is to show how many of RStudio’s features can be used during the process to speed the task along. We will postpone for now an example of the “development” aspect of RStudio.
The data set we look at here comes from a colleague, and contains records from a psychology experiment on a colony of naked mole rats. The experimenter is interested in both the behavior of each naked mole rat in time and the social aspect of the colony as a whole.
Each rat wears an RFID chip that allows the researcher to track its motion. The experiment consists of 15 chambers (bubbles) in a linear arrangement separated by 14 tubes. Each tube has a gate with a sensor. When a mole rat passes through the tube, the time and gate are recorded. Unfortunately, gates can be missed, and the recording device can erroneously replicate values, so the raw data must be cleaned up.
This data comes to us in rich-text format (rtf). This quasi text-based format is a bit unusual for data transfer but presumably is used by the recording apparatus. We will see that this format has some idiosyncrasies that will require us to work a little harder than we might normally do to read data into an RStudio session, but don’t worry, RStudio is up to the task.
Our first step is to copy the file into a directory named NMR. We are performing this analysis using the
desktop version, so we simply copy the file the usual way after making a new
directory. Had we been working through a server, we could have uploaded the
file into a new directory using first the New
Folder toolbar button, then the Upload toolbar button of the Files component.
Using Projects
To organize our work, we set up a new project (see the section Organizing Activities with Projects). RStudio allows us to compartmentalize our
work into projects that have separate global workspaces and associated
files and integrate seamlessly with version control systems. We easily
navigate between projects using a selector (a combobox) in the main
toolbar located in the upper-right corner. The same selector has an option
to create a New Project..., which we
choose. To create a new project, one fills in a project name and location,
and if available, one can specify if version control is to be used.
When the project is created, the working directory is set. The title
bar of the Console pane is updated, as
are the contents of the Files
component, which lists the files and subdirectories in a given directory.
The Files pane resides by default in
the lower-right corner. If it isn’t showing, select its tab. In Figure 2-1, we see that our working directory contains our
data file and a bookkeeping file written when RStudio created the
package.
The Files browser pane is typical
of RStudio’s components. In addition to the main application toolbar, most
components come with their own toolbar. In this case, the toolbar has
buttons to add a new folder, delete selected files, etc. In addition, the
Files component adds a second toolbar
to facilitate the selection of files and navigation within
directories.
Reading in a Data File
Clicking on the data file name in the file browser opens up a system
text editor (Figure 2-2), allowing us to edit the
file. For many text-based files, the file will open in RStudio’s
source-code editor. However, the actual editor employed depends on the
extension and MIME type of the file. For rtf files,
the underlying operating system’s editor is used, which for Mac OS X is
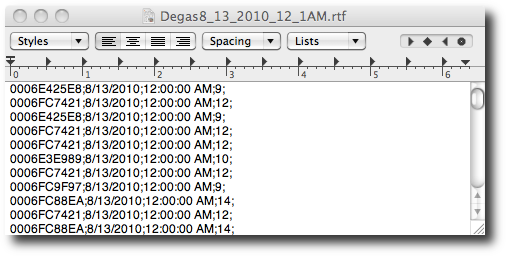
textedit. We can see that the data
appears to have one line per record, with the values separated by
semicolons. The fields are RFID, date, time, and gate number. This is
basically comma-separated-value (CSV) data with a nonstandard
separator.
However, although we rarely see rtf files, we
know the textedit program of Mac OS X
will likely render them using the markup for formatting, so perhaps there
are some markup commands that need to be removed. To investigate, we make
a copy of the data file, but store it instead with a
txt extension. The Files component makes it easy to perform basic
file operations such as this. To make a copy of a file, one selects the
checkbox next to the file and invokes the More
> Copy… menu item, as seen in Figure 2-3.
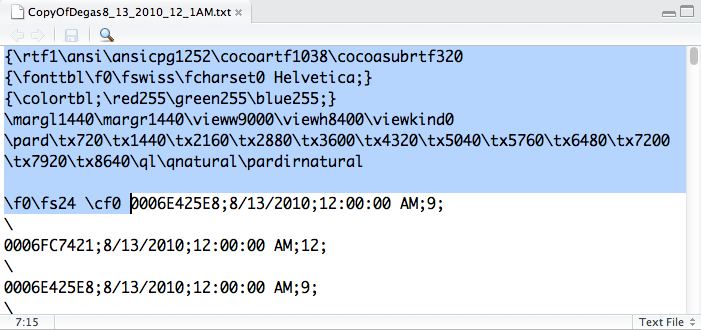
We change the extension to txt and our file list is updated. (In general this can be a really bad practice, especially for binary files, though in this case we know rtf files can be viewed as plain text.) The displayed contents of the directory may also be refreshed by clicking the terminus on the path indicated by the links to the right of the house icon in the secondary toolbar; or the refresh icon on the far right of the component’s main toolbar. Now, clicking on the txt file opens the file in RStudio’s source-code editor as a text file (Figure 2-4).
The editor’s status bar shows us the line and position of the cursor and, on the far right, that we are looking at a text file. We can now see that there is indeed a header (and, if we scroll down, a footer) wrapping our data. We highlight the header and then use the Delete key to remove this content from the file. We then scroll to the bottom of the file and remove a trailing brace. Afterwards, we click the Save toolbar button (the floppy-disk toolbar button, which is grayed out in the figure, as no changes have been made).
We now wish to read in the file using read.csv. RStudio provides an Import Dataset toolbar button under the Workspace component, which provides an interface
that will handle most csv data, such as that exported
from a spreadsheet. In this example though, we have a few idiosyncrasies
that prevent its use. (This is a deliberate choice to show off some of
RStudio’s other features.)
So we head on over to the Console
component to do the work. With the default pane arrangement the console is
located on the left side (the lower-left pane if the editor is open). In
R, one can’t avoid the console, and RStudio’s should look very familiar to
any R user.
Tab Completion
At the console we create the command to call the read.csv function directly. This requires us to
specify a few of its arguments, as we have a different separator, an odd
character every other line, and no header. We will use the tab completion
feature to assist us in filling in these values. This feature provides
completion candidates for many different settings, allowing us in this
case to recall quickly the names for lesser-used arguments.
First, we type read.csv in the
console. Then we press the Tab key to bring up the tab completion dialog
(Figure 2-5) for this function.
RStudio’s tab completion dialog for a function nicely displays its
arguments and a short description, gleaned from its help page (when
available). In this example we see the sep argument is what we need to specify a
semicolon for a separator, the header
argument to specify a non-default header, and comment.char to skip the lines starting with a
backslash.
The file name is the first argument. For file names (indicated by quotes), tab completion will fill in the file name, or, if more than one candidate is possible, provide a popup (Figure 2-6) to fill in the file. Here we type a left parentheses and double quote, and RStudio provides the matching values.
We press the Tab key again to select the proposed completion value using our modified text file, not the original. We then add a comma and again press the Tab key. When the prompt is in a function body, the tab completion will prompt for function arguments. After entering our values, we have this command to issue (see also Figure 2-7, where the command is shown in the console):
> x <- read.csv("CopyOfDegas8_13_2010_12_1AM.txt", sep=";",
+ header=FALSE, comment.char="\")Caution
The backslash argument for command.char is doubled, thereby escaping it.
Failing to do this, the parser will use the backslash to escape the
matching quote, getting the parser confused, as no matching quote will
be found. Pressing the Escape key will terminate the continuation prompt
so that the command can be fixed.
Workspace Component
The Workspace component lists the
objects in the project’s global workspace. In the default pane layout,
this component is in the upper-right pane along with the History component. If this pane isn’t raised, we
simply click on its tab (or perform the keyboard shortcut Ctrl-3) to do
so. After the data is read in, this component is updated to reflect the
new object, in this case one named x
(Figure 2-8). The associated icon for x shows it to be rectangular data. Clicking on
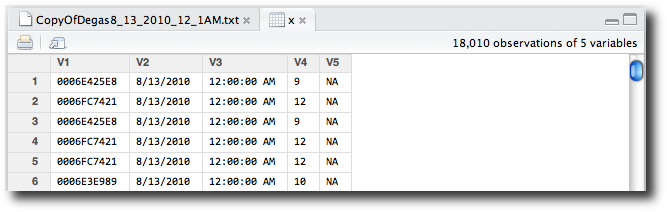
x’s row invokes the View function on x—in this case, opening the data viewer (Figure 2-9).
The data viewer shows us that we have an unnecessary fifth column of
NA values, and that our variable names
need improvement. Although the data viewer of RStudio does not yet support
editing, R has many ways to manipulate rectangular data at the command
line. For our two tasks we issue the following:
> x <- x[ , -5]
> names(x) <- c("RFID", "date", "time", "gate")The view of x in the code-editor
pane does not update from changes at the command line; rather, it is a
snapshot. The Workspace component does
reflect the current state of the variable, and reclicking on that will
refresh the view.
Using the Right Class to Store Data
The data is time-series data, but the date and time are read in
and stored by read.csv as factors,
not times. R has many different classes for working with time-series
data. In this case study we will look at two. The POSIXct class records time by the number of
seconds since the beginning of 1970 and is useful for storing times in a
data frame, such as x. We will use
the coercion function as.POSIXct for
this task. As this function isn’t part of our daily repertoire, we call
up its help page. Opening a help page can be done in the standard way:
?as.POSIXct (Figure 2-10).
Help pages are displayed in the Help pane, located by default in the
lower-right corner. RStudio’s help browser also has a search box on the
upper right of its main toolbar to locate a help page, or the page can
be opened with tab completion and the F1 key. Due to its web-technology
roots, RStudio easily leverages R’s HTML help system; pages appearing in
the Help pane have active
links.
After consulting the help page, we see that the format argument is needed. This specification
is described elsewhere, in the help page for the strptime function. Clicking on the provided
link opens that page, allowing us to figure out that the specification
needed to make our function call is:
> x$datetime <- paste(x$date, x$time) > x$time <- as.POSIXct(x$datetime, format="%m/%d/%Y %H:%M:%S")
Data Cleaning
At this point we have a data frame, x, storing all the information we have about the
colony of mole rats. However, the data set needs to be cleaned up, as
there are some repeated observations. We do this on a per-rat basis. R has
several ways to implement the split-apply-combine idiom, as it is one of
the most useful patterns for R users. The plyr package is widely used, but for this task
we use functions from base R. The split
function can be used to divide the data by the grouping variable RFID, returning a list whose components are the
records for the individual mole rats:
> l <- split(x, x$RFID)
The list, l, has a different
component for each mole rat. We can check to see if any two rows for a
mole rat are identical, using R’s convenient duplicated method. In addition, we add a bit of
time to to each time value, so that times recorded with the same second
are distinguished. R has several different means to apply a function to
pieces of an object. Below we use lapply to apply a function to each component of
the list l, returning a new list
l1 with the modified data:
> l1 <- lapply(l, function(x) {
+ trimmed <- x[duplicated(x),]
+ nr <- nrow(trimmed)
+ trimmed$time <- trimmed$time + seq_len(nr)/nr*(1/1000)
+ trimmed
+ })The data is recorded by gate, but the actual item of interest is the bubble (chamber) the mole rat is in at a given time. This information allows us to consider how social an animal is by looking at the time shared with others. We need to deduce this information from the data.
We do so by assuming that if the mole rat is in bubble 5, say, and
we record gate 5, then the mole rat moved to bubble 6. Or, if the
recording was gate 4, then the mole rat moved to bubble 4. (There are 15
bubbles and 14 gates, so gate i is between bubbles
i and i+1.) To create the bubble
count, we assume the mole rat moves immediately to the bubble after
crossing a gate. This ignores the possibility of the mole rat changing its
mind and never actually going to the next bubble. We will use a for loop to do this computation.
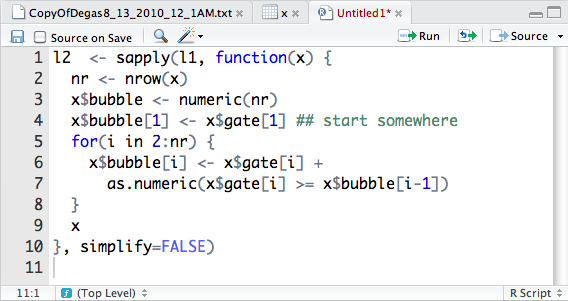
Using the Code Editor to Write R Scripts
The actual command we need for this computation is a bit long to type in correctly at the command line. We will instead use a script file so we can freely edit our commands. RStudio makes it easy to evaluate lines from a script file in the console. In addition, with the aid of syntax highlighting and automatic code formatting, we can quickly identify common errors before evaluation.
The “open a new R Script file”
action is proxied in several places: through the leftmost toolbar button
in the application toolbar, through the File >
New > R Script menu item, or through a keyboard shortcut
(Ctrl+Shift+N). However invoked, once done, a new untitled file appears in
the code-editor. In this new file we type in our commands, as shown in
Figure 2-11. The figure also shows how the code editor
component is used in many ways: to look at raw data sets, view rectangular
data objects from the workspace, and edit R commands.
With the commands typed in, we are ready to execute them. RStudio
allows several variations on how to send the contents of a file to the
console. In this case, we simply click on the Source toolbar button at the far right of the
pane’s toolbar to call source on the
active document.
Using Add-On Packages
Each component of the l2 list
contains records for a mole rat. The key variables are the times, stored
as POSIXct values and bubble. It will be more convenient to use
another of R’s date-time classes to represent the data, as then many
desirable methods will come along for free. Our data is an irregular time
series, as time is marked by mole rat events, not regular intervals on the
clock. The zoo package is designed for
such data, as one needs only ordered observations for the time
index.
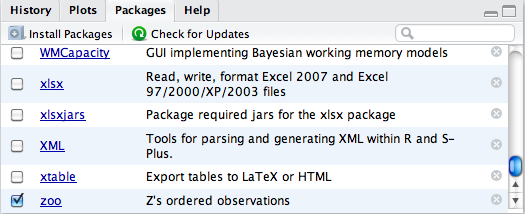
To convert our data into zoo
objects, we first need to load the package. RStudio makes working with
packages easy through the Packages
component, which for us appears in the lower-right pane. Loading or
unloading a package is as simple as checking the package’s accompanying
checkbox to indicate the desired state (Figure 2-12), where a check indicates the package is
loaded.
We had previously installed the zoo package, so it shows in the packages list.
Were that not the case, we could have quickly installed the package from
CRAN, along with any dependencies, using the dialog raised by clicking the
leftmost Install Packages toolbar
button in the pane’s toolbar.
To create a zoo object, we call
its same-named constructor. The first argument is the data; the second the
value to order by. We then merge the data into one zoo object. Here, we also use the na.locf function to carry the last bubble
forward to replace an NA when the data
is merged:
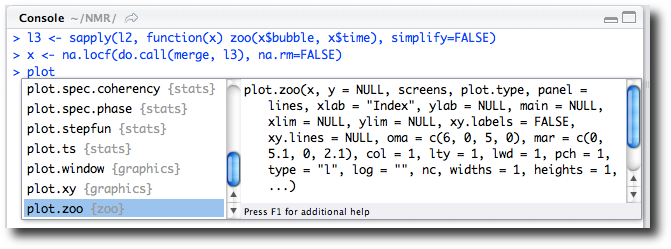
> l3 <- sapply(l2, function(x) zoo(x$bubble, x$time), simplify=FALSE) > x <- na.locf(do.call(merge, l3), na.rm=FALSE)
Graphics
One of the reasons we used a zoo
object is its convenient plot method.
We begin by making time series plots of the first five mole rats on the
same graphic. We can’t recall the specific arguments, so again let tab
completion (Figure 2-13) lead us to the correct
help page. In this case we type plot,
and the function completion shows us the various plot methods available. Scrolling through, we
find plot.zoo.
In the figure we see the plot.type argument for this plot method but
don’t recall the values to specify the graphic we desire. As instructed,
we press the F1 key to call up additional help in the help browser and
read that "single" is the desired
argument value.
After we issue the command:
> plot(x[, 1:5], plot.type="single")
the Plots component is raised,
showing the plot.
Command History
Noting that the individual paths are hard to distinguish once
they’ve crossed, we want to add colors to the graphic. The col argument is used for this. Rather than
retype the previous command, we can edit it. RStudio keeps a record of
previous commands. The up and down arrow shortcuts can be used to scroll
through our command history. For more complicated usage, we can use the
History component, which allows us to
browse the past commands and reissue them. We use the up arrow for this
case, then modify the col argument to
a simple value of 1:5, producing
Figure 2-14.
The plot is sized to fill the Plots pane, and
can be on the small side. (Unlike most interactive R use, where the plot
devices choose their size.) Often this is all that is needed, but in
this particular case we wish it to be bigger. The Zoom toolbar button of the Plots component’s toolbar will open the graph
in a larger window.
All Finished, for Now
At this point, with the help of RStudio, we have completed the data
preparation needed for subsequent analysis. We have a zoo object holding all the data (x) and a list of zoo objects (l3) storing data for individual rats. In the
process of this 30-minute analysis, we took advantage of most all of
RStudio’s key components: the Files
browser, tab completion, the text editor, the Help browser, the rectangular data viewer, the
Console, the Source code editor, the Packages browser, and the Plots viewer.