Chapter 2
Overview of Methods for Dealing with Missing Data
Regardless of the existence of missing data, the end result of any analysis is to make valid and efficient inferences about the population of interest. Neyman and Pearson 1933 established valid criteria for evaluating any statistical procedure. These criteria include having a small bias, where bias refers to the difference between the average sample estimate and its true value, and a small variance associated with the average sample estimate (efficiency). Bias and variance can be combined in a single measure called the mean square error (MSE) so that the bias, variance, and MSE describe the behavior of the estimate. Using these criteria, we discuss the various missing data methods that are available, each with its own strengths and limitations. This chapter provides a brief overview of the existing approaches and classifies them according to whether they remove observations with missing data, utilize all available data, or impute missing data. An excellent overview of missing data methods is provided by Schafer and Graham 2002.
2.1 Methods that Remove Observations
Many approaches simplify the missing data problem by discarding data. We discuss how most of these approaches may lead to biased results, although some approaches try to directly address this issue. Discarding data also reduces the sample size, which in turn reduces efficiency and leads to larger standard errors for the parameters of interest.
2.1.1 Complete-Case Methods
As the name suggests, the complete-case method (also known as list-wise deletion) makes use of cases with complete data on all variables of interest in the analysis. Cases with incomplete data on one or more variables are discarded. This approach is easiest to implement since standard full data analysis can be applied without modification, and is the default in many statistical software packages (i.e., cases with any missing values are automatically ignored). The disadvantages include loss of precision due to a smaller sample size and bias in complete-data summaries when the cases with 17 missing data differ systematically from the completely observed cases (i.e., when data are not missing completely at random).
In practice, when the loss of precision and the bias are small, we may opt for this approach for its simplicity. Unfortunately, the loss of precision and the bias are difficult to quantify since both depend not only on the fraction of complete cases but also on the extent to which complete and incomplete cases differ on the parameters of interest. So there is no universal rule of thumb, such as a certain percentage of missing data, for deciding whether this is an appropriate method.
2.1.2 Weighted Complete-Case Methods
The earlier complete-case strategy can potentially lead to biased summaries because the sample of observed cases may not be representative of the full sample. Another strategy reweights the sample to make it more representative. For example, if the response rate is twice as likely among men as women, data from each man in the sample could receive a weight of 2 in order to make the data more representative. This strategy is commonly used in sample surveys, especially in the case of unit nonresponse, where all the survey items are missing for cases in the sample that did not participate. For example, if only one variable is missing, this method first creates a model to predict nonresponse in one variable as a function of the rest of the variables in the data. The inverse of the predicted probabilities of response from this model could then be used as survey weights to make the complete-case sample more representative. If more than one variable is missing, this method becomes more complicated, and there will be problems with standard errors if predicted response probabilities are close to 0.
We give an example to illustrate this method more formally. Let Y be the outcome of interest and R be the indicator of whether Y was observed. We also let X denote the set of variables that were observed for both respondents and nonrespondents. Suppose that the MAR (missing at random) assumption holds, that is, Pr (R|X, Y) = Pr (R|X). Then the response probability of the ith subject is denoted by
If π(Xi) values are known for all n subjects in the sample, we could use
to estimate the marginal mean of Y, which is the so-called inverse probability weighted estimator (Cassel et al., 1983; Skinner and D'arrigo, 2011. If the parameter θ is interest, which satisfies E[g(Y, X; θ)] = 0, we could solve the following estimating equation to find an estimate of θ:
In many practical applications, the response probability π(X) is unknown. In this case, we may assume a parametric model for it, that is, π(X) is known up to some parameters. Or we can estimate π(X) nonparametrically if X does not contain many variables. In some applications, we may group the values of π(X) into four to six categories and create a new variable C, which is used to adjust weights in the analysis. This might be required because directly weighting by [π(X)]−1 places more reliance on correct model specification of the regression of R on X than adjusting for C (Little and Rubin, 1987).
2.1.3 Removing Variables with Large Amounts of Missing Values
This approach drops variables from the analysis that have a large proportion of missing values. This is not recommended since it may exclude important variables in the regression model, required for causal interpretation, and may lead to bias and unnecessarily large standard errors. For important variables with more than trivial amounts of missing data, the explicit model-based methods in the next sections are suggested.
2.2 Methods that Utilize all Available Data
In contrast to complete-case methods, available case methods (also known as pairwise deletion methods) utilize information from both completely and partially observed cases. In multivariate regression, cases with incomplete data on certain variables can still provide information about the relationship between the outcome and other observed variables. For example, in repeated measures data, cases with incomplete data at one time point can still provide valid information about the relationship between variables at other time points. Mixed effects models are therefore a popular choice for dealing with longitudinal data with missing outcomes. Because the available case methods make use of more information than the complete-case methods, they are generally better at correcting for bias when data are MAR than the complete-case methods.
A major disadvantage of this approach is that different analyses would use different subsets of the data, depending on the variables used and their missing data patterns, which may not provide consistent inference. And similar to complete-data methods, there will be a loss of efficiency from discarding the partially observed variables, and the available case summaries will be biased when the cases with missing data differ systematically from the observed cases.
2.2.1 Maximum Likelihood
Many procedures arise from defining a model for the variables with missing values and making statistical inferences based on maximum likelihood (ML) methods. The fundamental idea behind the ML methods is conveyed by their name: find the values of the parameters that are most probable, or most likely, for the data that have actually been observed.
Denote Dobs as the observed data, which have the probability density p(Dobs|θ). Here θ is a set of parameters of interest. Then, the likelihood function L(θ|Dobs) is equal to p(Dobs|θ), that is, L(θ|x) = p(Dobs|θ), and is thought of as a function of θ, where the data Dobs are fixed. The log-likelihood l(θ|Dobs) is equal to log (L(θ|x) and is, in general, easier to be maximized.
The next step is to deduce a numerical value for θ using our knowledge of L(θ|x). The ML principle says we should choose an estimate of θ that maximizes the log-likelihood function. In other words, we choose a value of the parameter that best explains the observed data, and this value is called the ML estimate of θ.
Finding the maximum can be easy or hard, depending on the form of p(Dobs|θ). For the normal (Gaussian) distribution, θ consists of the mean μ and variance σ2, and we can find the derivative of l(θ|Dobs), set it equal to zero, and solve directly for μ and σ2. But for most problems, these analytic expressions are hard to come by, and we will have to use more elaborate techniques. One might use the Newton–Raphson or quasi-Newton algorithms to maximize directly the likelihood of the observed data. One of the most popular techniques for maximizing the likelihood function is the expectation–maximization (EM) algorithm of Dempster et al. 1977. We briefly discuss the EM algorithm in Section 2.2.1.1 and readers may find the theoretical details in Little and Rubin (1987).
2.2.1.1 EM Algorithm
The EM algorithm is a general method of finding the ML estimate of the parameters of an underlying distribution from a given data set when the data are incomplete or have missing values.
The EM algorithm has two main applications: First, to find the ML estimates when data truly have missing values (e.g., patients lost to follow-up); Second, to find the ML estimates when the likelihood is analytically intractable and difficult to be optimized, but the likelihood can be simplified by assuming that there exist additional, but missing parameters. The algorithm can be applied whether the missing data are ignorable or not by including a missing data model in the likelihood.
The EM algorithm is essentially an iterative optimization algorithm that will, under certain conditions, converge to parameter values at a local maximum of the likelihood function. Let Dmis be the missing data. Then, the full data Dcom = {Dobs, Dmis}. In general, the full data log-likelihood will have an easily defined, analytically solvable maximum, but maximization of the observed-data log-likelihood l(θ|Yobs) may have no analytic solution.
The EM algorithm approach is a procedure that iterates between two steps: an expectation step (E step) and a maximization step (M step). The E step calculates the expected value of the full data log-likelihood, given the observed data and current estimated parameters. The M step performs maximization on the expectation of the full data log-likelihood, computed in the E step above. The steps are usually easy to construct programming, and implementation of the EM algorithm is straightforward using standard statistical software packages.
2.3 Methods that Impute Missing Values
In contrast to methods that remove observations with missing values, imputation methods impute or “fill in” missing values. Perhaps because of long-standing taboos against “making up” data, it has taken some time for imputation methods to gain general favor among the research community despite being as sound as other acceptable approaches. The key realization behind imputation is that, although the missing values are not observed, information about these values may still be extracted from the other observed variables.
This section will consider methods that impute, or fill in, missing values. These methods can be implemented by imputing one value for each missing item (single imputation), or by imputing more than one value to allow for appropriate assessment of imputation uncertainty (multiple imputation). In general, anytime a single imputation strategy is used, standard errors of estimates of parameters will be underestimated since the resulting data are treated as a complete sample, ignoring the consequences of imputation. In contrast, multiple imputation strategies take into account the uncertainty about the imputed missing values, and are therefore preferable to single imputation. The single and multiple imputation strategies are discussed in Sections 2.3.1 and 2.3.2, respectively.
2.3.1 Single Imputation Methods
2.3.1.1 Unconditional Mean Imputation
A popular approach replaces each missing value with the unconditional mean of the observed values for a particular variable. This is a single imputation approach and can therefore lead to underestimating standard errors. Additionally, this method distorts relationships between variables since it pushes the correlations between variables toward zero (Gelman and Hill, 2006).
2.3.1.2 Conditional Mean Imputation
For data sets with few missing variables, conditional mean imputation (also known as regression imputation) is an improvement on the unconditional mean approach since it replaces each missing value with the conditional mean of the variable based on other fully observed variables in the data set. If there is only one variable with missing values, say Y, then a regression model for predicting Y from the other variables X = (X1, ..., Xk) ' can be used to impute Y. The first step fits the model to the cases where Y is observed. The second step plugs in the X values for the nonrespondents into the regression equation, obtaining predicted values ![]() for the missing values of Y. This is a single imputation approach and can therefore lead to underestimating standard errors, although this approach is acceptable for some problems if the standard errors are corrected (Schafer and Schenker, 2000). However, it is not recommended for analyses of covariances and correlations since it distorts the strength of the relationship between X and Y. This approach is especially problematic when one is interested in the details of the distribution (Little and Rubin, 1987). For example, an approach that imputes missing incomes using conditional means tends to underestimate the percentage of cases in poverty.
for the missing values of Y. This is a single imputation approach and can therefore lead to underestimating standard errors, although this approach is acceptable for some problems if the standard errors are corrected (Schafer and Schenker, 2000). However, it is not recommended for analyses of covariances and correlations since it distorts the strength of the relationship between X and Y. This approach is especially problematic when one is interested in the details of the distribution (Little and Rubin, 1987). For example, an approach that imputes missing incomes using conditional means tends to underestimate the percentage of cases in poverty.
2.3.1.3 Last Observation Carried Forward (LOCF)
For longitudinal studies, this approach imputes the missing value at one time point with its value at the previous time point, or uses the last observation carried forward. Both mean imputation and LOCF are not recommended since they can potentially induce bias as well as underestimate variability (Sasha and Jones, 2009).
2.3.1.4 Substitution of Related Observations
Other methods may utilize data from related observations. For example, suppose that we are missing the education level of some of the fathers of children. We might fill in these values using the mothers' report of the fathers' education level. This may seem plausible, but it introduces measurement error, especially if there is a reason to believe that the reporting subjects may misrepresent the measurements of the people about whom they are providing information. As another example, if a household cannot be contacted, we might substitute their missing values with values from a household in the same neighborhood.
2.3.1.5 Hot Deck Imputation
This approach selects a value at random from the pool of “similar” complete cases, and the missing value is replaced by the selected value. An advantage of this approach is that it does not require careful modeling to develop the selection criteria for imputing the value, although bias can still be introduced. Note that this approach can serve as a multiple imputation method by selecting multiple values at random from the pool.
2.3.1.6 Missing Indicator Method
Some ad hoc approaches seem to solve the missing data problem by retaining the data, but in reality, merely redefine the parameters of the population. Consider a linear regression of Y on X, where X has some missing values. Suppose we recode all missing values of X to a common number and create a binary indicator R, which is equal to 1 when X is missing and 0 otherwise. In our original model, E(Y|X) = β0 + β1X, where β0 and β1 represent the intercept and the slope of the full population, respectively. Adjusting for missing indicator R, the model becomes E(Y|X) = β0 + β1X + β2R, where β0 and β1, respectively, become the intercept and slope among respondents and β0 + β2 becomes the intercept for nonrespondents. In particular, this method forces the slope to be the same across groups with and without missing X, which will likely lead to biased coefficient estimates.
Adding an interaction in the model can help lessen this bias, that is, replace the term β1X with β1X(1 − R), and this approach leads to estimates similar to the complete-case estimates. But in general, these approaches are not recommended because of the potential bias they induce in their attempt to estimate population-level summaries (Jones, 1996; Greenland and Finkle, 1995).
For categorical variables with missing values, this approach creates an additional category for missing values. In the regression example above, if X is a nominal variable with values 1, ..., k, then treating the missing value as category k + 1 merely redefines categories 1, ..., k to apply to respondents only. This approach can lead to biased results, even if the data are missing completely at random (MCAR).
2.3.2 Multiple Imputation
While imputation may solve the missing data problem, naive or unprincipled imputation methods may create more problems than they solve, by distorting estimates and standard errors. Rubin (1987) addressed the question of how to obtain valid inference from imputed data by using multiple imputation, a technique where missing data are replaced by m values drawn from the posterior predictive distribution of the missing data, resulting in m complete data sets. Standard full data methods are then used on each of the m complete data sets to estimate the parameters of interest, resulting in m estimated parameters and their corresponding standard errors. These estimates are then combined, or averaged, to produce estimates and confidence intervals that incorporate the missing data uncertainty. This averaging will, in general, lower the variance of the combined estimate. In particular, this method recognizes two sources of uncertainty: the uncertainty caused by sampling the subjects from a source population, and the uncertainty in estimating underlying distributions of the variables with missing values. A simple method is used to combine both sources of uncertainty, resulting in a single corrected standard error of the estimated association.
The multiple imputation method was originally developed in the setting of large public-use data sets from sample surveys and censuses. However, with advances in computational methods and software for creating multiple imputations, the technique has become attractive to researchers in most fields where missing data cause a problem, as documented in Schafer (1997b).
Software most readily available to implement multiple imputation assumes that the missing data are MAR, although the paradigm certainly does not require MAR nonresponse. In principle, multiple imputations can be created in any kind of model for the missing data mechanism, and resulting data will be valid under that mechanism (Rubin, 1987). But methods that assume MAR have the advantage of avoiding explicit modeling of nonresponse, although unfortunately the observed data cannot provide enough information on the validity of the MAR assumption.
Usually the imputed values are drawn from an imputation model. If only one variable is subject to missingness, it is easy to construct the imputation model. For example, we can use logistic regression to model a binary variable, and a linear Gaussian model for a continuous variable. If there are multiple variables subject to missingness, the imputation model can be constructed jointly or conditionally.
The joint modeling approach assumes that given the other observed variables, the variables subject to missingness follow a multivariate normal distribution. However, in practice, the variables subject to missingness may involve different types such as binary, categorical, and ordinal variables. In this case, it would be computationally implausible to simulate the posterior predictive distribution simultaneously for all the missing variables. The multiple imputation with chained equations is proposed. The idea of chained equations is to update the missing variables one by one, based on a series of full conditional distributions, as proposed by van Buuren and Groothuis-Oudshoorn (2011).
Denote Y = (Y1, ..., Yp)′ the set of p variables in the data set that are subject to missingness, which include both response variables and covariates. Let Y−j = (Y1, ..., Yj−1, Yj+1, ..., Yp)′ be the collection of all variables in Y but Yj. The model assumptions are made for each variable Yj fully conditional on all other variables Y−j:
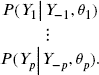
This model framework is called “chained equations”. It specifies p univariate regression models, and avoids the hassle of jointly model the multivariate distributions for all the missing variables.
With a sample of size n, the observed data on Yj is a vector of observations from n subjects, denoted by yj. Hence we can partition yj into the missing part ![]() and the observed part
and the observed part ![]() , indicating a pool of subjects with observed or missing Yj. It should be noted that this is a partition of subjects for each variable, whereas the partition of variables for a subject will be introduced in later chapters. The iteration procedures are described as follows:
, indicating a pool of subjects with observed or missing Yj. It should be noted that this is a partition of subjects for each variable, whereas the partition of variables for a subject will be introduced in later chapters. The iteration procedures are described as follows:

where ![]() is the imputed data vector of yj after the tth iteration. The draws for the parameters are just a full Bayesian regression analysis. Note that for drawing
is the imputed data vector of yj after the tth iteration. The draws for the parameters are just a full Bayesian regression analysis. Note that for drawing ![]() , the condition is on
, the condition is on ![]() rather than
rather than ![]() . In other words, only subjects with observed response (Yj) are included in the regression; the covariates (Y−j) are all imputed based on the previous iteration. The draws for the missing data are performed sequentially for each variable on the subjects with missing values, which is based on the p univariate regression models. This procedure has been implemented in the R package mice.
. In other words, only subjects with observed response (Yj) are included in the regression; the covariates (Y−j) are all imputed based on the previous iteration. The draws for the missing data are performed sequentially for each variable on the subjects with missing values, which is based on the p univariate regression models. This procedure has been implemented in the R package mice.
In practice, the chained equation approach can be briefly broken down into the following steps, and we will illustrate these with examples in the later chapters.
- Examine the missing data patterns and identify key variables with missing values that require imputation. Decide on the sequence of the imputation, usually based on amount of missingness and quality of the variables used to impute them.
- Initialize all missing values with a simple imputation, such as mean imputation or last-value-carry-forward.
- Construct an imputation model (typically a regression model) for the first variable (Y1) requiring imputation. For example, if Y1 is a binary variable, a logistic regression model may be constructed with Y1 being the dependent variable, and other variables as covariates. For other types of variables, linear, Poisson or other generalized linear regression models may be employed.
- The missing values for Y1 are then replaced with predictions (imputations), using either predictive mean matching method or direct sampling from predictive posterior distributions. Note the latter method may require Gibbs sampling for intractable distributions. When Y1 is used in subsequent imputation models for other variables, the observed values and imputed values for Y1 will be combined to form a complete data vector.
- Steps 3 and 4 are repeated for each variable that has missing values, Y2, ..., Yp. The cycling through each of the variables constitutes one iteration. At the end of each iteration all of the missing values have been replaced with imputed values from regressions that are assumed to reflect relationships among these variables. The number of iterations to be performed can be decided by the researchers. Though it is suggested that 10 iterations are usually sufficient, nowadays with the increasing computing power, we recommend performing iterations many more times to ensure convergence of the algorithm. The imputed data set in the last iteration is taken to be one imputed data set.
- Iterate steps 2 through 5 to obtain multiple imputed datasets. The number of imputations should depend on the missing information of the data set. In most cases, 10-20 imputations would be sufficient.
- With each imputed complete dataset, we then proceed with parameter estimation. The results are pooled together over all imputed datasets for the final inference.
The chained equation is able to handle flexible type of missing variables, and is usually simple to implement. Although the convergence of the algorithm is not proved rigorously, its performance in practice is stable in most circumstances, as long as the missing data proportion is not too high. One important limitation of the chained equation approach is that there is no guarantee that the imputed sample is indeed drawn from the posterior predicted distribution because there may not exist a joint distribution that yields the specified conditional distributions.
2.4 Bayesian Methods
Bayesian statistical conclusions about the parameter θ or missing data Dmis are made in terms of probability statements that are conditional on the observed value Dobs, or p(θ|Dobs) and p(Dmis|Dobs). It is exactly this conditioning on the observed data that makes Bayesian inference different from the commonly used statistical inference described in most articles and textbooks, which is based on an evaluation of the procedure used to estimate θ or Dmis over the distribution of possible Dobs values conditional on the true unknown value of θ. Note that in simple settings, results from both methods can be quite similar.
Bayesian methods make no distinction between the missing data and the parameters since both are uncertain. Focus is placed on the joint distribution of the missing data and parameters conditional on observed data, and the typical setup for the joint distribution includes a prior distribution for the parameters, a joint model for all the data (observed and missing), and a model for the missing data process (unless the missing data mechanism is ignorable, in which case this last model is not necessary).
Missing variables are sampled from their conditional distribution via different methods such as the Gibbs sampler. There is an imputation stage where simulated draws from the posterior predictive distribution of unobserved values Dmis conditional on observed values Dobs are imputed. Then there is a draw from the posterior distribution of the parameter θ. Inference then proceeds by averaging over the distribution of missing values. Fully Bayesian methods with missing variables only involve incorporating an extra step in the Gibbs sampler, compared with the case of having no missing values in the data. Therefore, Bayesian methods do not require additional techniques for inference to accommodate missing data. For this reason, Bayesian methods are quite powerful.
