9.9 Exercises on Chapter 9
1. Find the value of  by crude Monte Carlo integration using a sample size of n=10 values from a uniform distribution U(0, 1) taken from tables of random numbers [use, e.g. groups of random digits from Lindley and Scott (1995, Table 27) or Neave (1978, Table 8.1)]. Repeat the experiment ten times and compute the overall mean and the sample standard deviation of the values you obtain. What is the theoretical value of the population standard deviation and how does the value you obtained compare with it?
by crude Monte Carlo integration using a sample size of n=10 values from a uniform distribution U(0, 1) taken from tables of random numbers [use, e.g. groups of random digits from Lindley and Scott (1995, Table 27) or Neave (1978, Table 8.1)]. Repeat the experiment ten times and compute the overall mean and the sample standard deviation of the values you obtain. What is the theoretical value of the population standard deviation and how does the value you obtained compare with it?
2. Suppose that, in a Markov chain with just two states, the probabilities of going from state i to state j in one time unit are given by the entries of the matrix

in which i represents the row and j the column. Show that the probability of getting from state i to state j in t time units is given by the tth power of the matrix and that
and that

Deduce that, irrespective of the state the chain started in, after a long time it will be in the first state with probability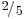 and in the second state with probabi-lity
and in the second state with probabi-lity  .
.
in which i represents the row and j the column. Show that the probability of getting from state i to state j in t time units is given by the tth power of the matrix
Deduce that, irrespective of the state the chain started in, after a long time it will be in the first state with probability
3. Smith (1969, Section 21.10) quotes an example on genetic linkage in which we have observations 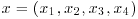 with cell probabilities
with cell probabilities

The values quoted are x1=461, x2=130, x3=161 and x4=515. Divide x1 into y0 and y1 and x4 into y4 and y5 to produce augmented data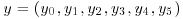 and use the EM algorithm to estimate η.
and use the EM algorithm to estimate η.
The values quoted are x1=461, x2=130, x3=161 and x4=515. Divide x1 into y0 and y1 and x4 into y4 and y5 to produce augmented data
4. Dempster et al. (1977) define a generalized EM algorithm (abbreviated as a GEM algorithm) as one in which 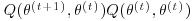 . Give reasons for believing that GEM algorithms converge to the posterior mode.
. Give reasons for believing that GEM algorithms converge to the posterior mode.
5. In question 16 in Chapter 2, we supposed that the results of a certain test were known, on the basis of general theory, to be normally distributed about the same mean μ with the same variance  , neither of which is known. In that question, we went on to suppose that your prior beliefs about
, neither of which is known. In that question, we went on to suppose that your prior beliefs about  could be represented by a normal/chi-squared distribution with
could be represented by a normal/chi-squared distribution with
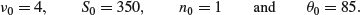
Find a semi-conjugate prior which has marginal distributions that are close to the marginal distributions of the normal/chi-squared prior but is such that the mean and variance are independent a priori. Now suppose as previously that 100 observations are obtained from the population with mean 89 and sample variance s2=30. Find the posterior distribution of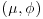 . Compare the posterior mean obtained by the EM algorithm with that obtained from the fully conjugate prior.
. Compare the posterior mean obtained by the EM algorithm with that obtained from the fully conjugate prior.
Find a semi-conjugate prior which has marginal distributions that are close to the marginal distributions of the normal/chi-squared prior but is such that the mean and variance are independent a priori. Now suppose as previously that 100 observations are obtained from the population with mean 89 and sample variance s2=30. Find the posterior distribution of
6. A textile company weaves a fabric on a large number of looms. Four looms selected at random from those available, and four observations of the tensile strength of fabric woven on each of these looms are available (there is no significance to the order of the observations from each of the looms), and the resulting data are as follows:
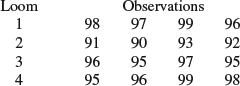
Estimate the means for each of the looms, the overall mean, the variance of observations from the same loom, and the variance of means from different looms in the population.

Estimate the means for each of the looms, the overall mean, the variance of observations from the same loom, and the variance of means from different looms in the population.
7. Write computer programs in C++ equivalent to the programs in  in this chapter.
in this chapter.
8. Use the data augmentation algorithm to estimate the posterior density of the parameter η in the linkage model in question 3.
9. Suppose that 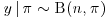 and
and  where n is a Poisson variable of mean λ as opposed to being fixed as in Section 9.4. Use the Gibbs sampler (chained data augmentation) to find the unconditional distribution of n in the case where
where n is a Poisson variable of mean λ as opposed to being fixed as in Section 9.4. Use the Gibbs sampler (chained data augmentation) to find the unconditional distribution of n in the case where 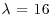 .
.  and
and 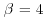 (cf. Casella and George, 1992).
(cf. Casella and George, 1992).
10. Find the mean and variance of the posterior distribution of θ for the data in question 5 mentioned earlier using the prior you derived in answer to that question by means of the Gibbs sampler (chained data augmentation).
11. The following data represent the weights of r=30 young rats measured weekly for n=5 weeks as quoted by Gelfand et al. (1990), Tanner (1996, Table 1.3 and Section 6.2.1), Carlin and Louis (2000, Example 5.6):
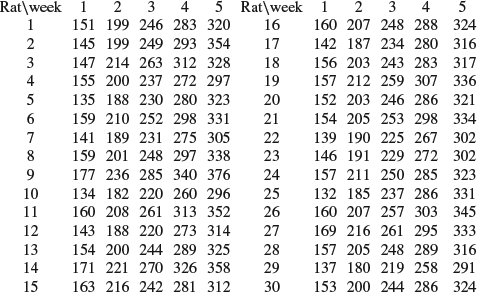

The weight of the ith rat in week j is denoted xij and we suppose that weight growth is linear, that is,

but that the slope and intercept vary from rat to rat. We further suppose that 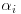 and
and 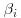 have a bivariate normal distribution, so that
have a bivariate normal distribution, so that
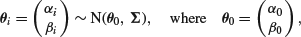
and thus we have a random effects model. At the third stage, we suppose that


where we have used the notation 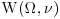 for the Wishart distribution for a random
for the Wishart distribution for a random 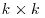 symmetric positive definite matrix
symmetric positive definite matrix 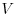 , which has density
, which has density
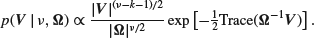
Methods of sampling from this distribution are described in Odell and Feiveson (1966), Kennedy and Gentle (1990, Section 6.5.10) and Gelfand et al. (1990). [This example was omitted from the main text because we have avoided use of the Wishart distribution elsewhere in the book. A slightly simpler model in which  is assumed to be diagonal is to be found as the example ‘Rats’ distributed with WinBUGS.]
is assumed to be diagonal is to be found as the example ‘Rats’ distributed with WinBUGS.]
Explain in detail how you would use the Gibbs sampler to estimate the posterior distributions of  and
and 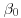 , and if possible carry out this procedure.
, and if possible carry out this procedure.
12. Use the Metropolis–Hastings algorithm to estimate the posterior density of the parameter η in the linkage model in Sections 9.2 and 9.3 using candidate values generated from a uniform distribution on (0, 1) [cf. Tanner (1996, Section 6.5.2)].
13. Write a WinBUGS program to analyze the data on wheat yield considered towards the end of Section 2.13 and in Section 9.3.
14. In bioassays, the response may vary with a covariate termed the dose. A typical example involving a binary response is given in the following table, where R is the number of beetles killed after 5 hours of exposure to gaseous carbon disulphide at various concentrations (data from Bliss, 1935, quoted by Dobson, 2002, Example 7.3.1).
Fit a logistic regression model and plot the proportion killed against dose and the fitted line.
| Dose xi | Number of | Number |
| (log10CS2mgl–2) | insects, ni | killed, ri |
| 1.6907 | 59 | 6 |
| 1.7242 | 60 | 13 |
| 1.7552 | 62 | 18 |
| 1.7842 | 56 | 28 |
| 1.8113 | 63 | 52 |
| 1.8369 | 59 | 53 |
| 1.8610 | 62 | 61 |
| 1.8839 | 60 | 60 |
Fit a logistic regression model and plot the proportion killed against dose and the fitted line.
1. Sometimes called a jumping distribution.
..................Content has been hidden....................
You can't read the all page of ebook, please click here login for view all page.
