Recipe 5-1. Aggregate data on a single key
Recipe 5-2. Aggregate data on multiple keys
Recipe 5-3. Create a contingency table
Recipe 5-4. Perform joining operations on two DataFrames
Recipe 5-5. Vertically stack two DataFrames
Recipe 5-6. Horizontally stack two DataFrames
Recipe 5-7. Perform missing value imputation
Recipe 5-1. Aggregate Data on a Single Key
Problem
You want to perform data aggregation on a DataFrame, grouped on a single key.
Solution
We perform data aggregation to observe summary of data.
Mean value of the number of accepted and rejected students
Mean value of students gender-wise who have applied for admission
The average frequency of application gender-wise
mean()
count()
sum()
min()
max()
etc ….
How It Works
Step 5-1-1. Reading the Data from MySQL
Step 5-1-2. Calculating the Required Means
Step 5-1-3. Grouping by Gender
Step 5-1-4. Finding the Average Frequency of Application by Gender
Recipe 5-2. Aggregate Data on Multiple Keys
Problem
You want to perform data aggregation on a DataFrame, grouped on multiple keys.
Solution
Group the data on the admit and gender columns and find the mean of applications to the college.
Group the data on the admit and department columns and find the mean of applications to the college.
How It Works
Step 5-2-1. Grouping the Data on Gender and Finding the Mean of Applications
Step 5-2-2. Grouping the Data on Department and Finding the Mean of Applications
Recipe 5-3. Create a Contingency Table
Problem
You want to create a contingency table.
Solution
Contingency tables are also known as cross tabulations. They show the pairwise frequency of the given columns.

Contingency table for the restaurant survey
We have to make a contingency table using the data in Figure 5-1.
How It Works
Step 5-3-1. Reading Restaurant Survey Data from MongoDB
Step 5-3-2. Creating a Contingency Table
We have our contingency table.
Recipe 5-4. Perform Joining Operations on Two DataFrames
Problem
You want to perform a join operation on two DataFrames.
Solution
Inner join
Left outer join
Right outer join
Full outer join

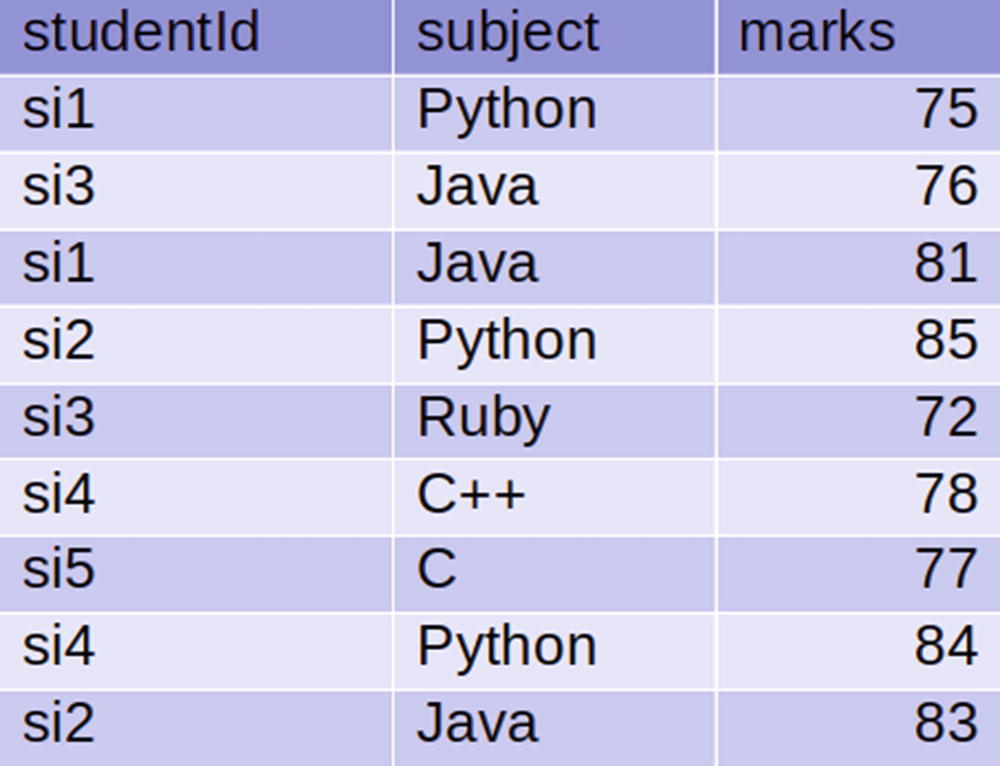
The students table
Read the students and subjects tables from the Cassandra database.
Perform an inner join on the DataFrames.
Perform an left outer join on the DataFrames.
Perform a right outer join on the DataFrames.
Perform a full outer join on the DataFrames.
In order to join the two DataFrames, PySparkSQL provides a join() function, which works on the DataFrames.
The join() function takes three arguments. The first argument, called other, is the other DataFrame. The second argument, called on, defines the key columns on which we want to perform joining operations. The third argument, called how, defines which join type to perform.
How It Works
Let’s explore this recipe in a step-by-step fashion.
Step 5-4-1. Reading Student and Subject Data Tables from a Cassandra Database
We need to read the students data in the pysparksqlbook keyspace of the students table. Similarly, we need to read the subjects data in the same keyspace, but in a Cassandra table called subjects.
We have dropped the unwanted column.
Step 5-4-2. Performing an Inner Join on DataFrames
At this moment, we have two DataFrames called studentsDf and subjectsDf. We have to perform inner joins on these DataFrames. We have a studentid column that’s common in both DataFrames. We are going to perform an inner join on that studentid column.
Let’s explore the argument of the join() function. The first argument is subjectsDf, which is the DataFrame that’s getting joined with the studentsDf DataFrame. The second argument is studentsDf.studentid == subjectsDf.studentid, which indicates the condition of joining. The last argument tells us that we have to perform an inner join.
In the output DataFrame innerDf, we can observe that in the studentid column, we see only si1, si2, si3, and si4 .
Step 5-4-3. Performing a Left Outer Join on DataFrames
In this step of recipe, we are going to perform the left outer join on the DataFrames. In a left outer join, each key for the first DataFrame and matched keys from the second DataFrame will be in the result.
The studentid column in the resulting DataFrame shows si1, si2, si3, si4, and si6 .
Step 5-4-4. Performing a Right Outer Join on DataFrames
The result of the right outer join is rightOuterDf .
Step 5-4-5. Performing a Full Outer Join on DataFrames
Recipe 5-5. Vertically Stack Two DataFrames
Problem
You want to perform vertical stacking on two DataFrames.
Solution

Vertically stacking DataFrames
Figure 5-4 depicts the vertical stacking of two DataFrames. On the left, we can see two DataFrames called dfOne and dfTwo. DataFrame dfOne consists of three columns and four rows. Similarly, DataFrame dfTwo also has three row and three columns. The right side of Figure 5-4 displays the DataFrame that’s been created by vertically stacking the dfOne and dfTwo DataFrames.
We have to read two tables—firstverticaltable and secondverticaltable—from the PostgreSQL database and create DataFrames.
We have to vertically stack the newly created DataFrames.
We can perform vertical stacking using the union() function . The union() function takes only one input, and that is another DataFrame. This function is equivalent to UNION ALL in SQL. The resultant DataFrame of the union() function might have duplicate records. It is therefore suggested that we use the distinct() function on the result of a union() function .
How It Works
The first step is to read the data tables from PostgreSQL and create the DataFrames.
Step 5-5-1. Reading Tables from PostgreSQL
Note
In order to connect with the PostgreSQL database, we have to start a PySpark shell with the following options:
$pyspark --driver-class-path ~/.ivy2/jars/org.postgresql_postgresql-42.2.4.jar --packages org.postgresql:postgresql:42.2.4
Step 5-5-2. Performing Vertical Stacking of Newly Created DataFrames
Finally, we have a vertically stacked DataFrame, named vstackedDf .
Recipe 5-6. Horizontally Stack Two DataFrames
Problem
You want to horizontally stack two DataFrames.
Solution

Horizontally stacking DataFrames
We can observe from Figure 5-5 that horizontal stacking means putting DataFrames side by side. It is not easy in PySparkSQL, as there is no dedicated API for this task.
Since there is no dedicated API for horizontal stacking, we have to use other API to get the job done. How do we perform horizontal stacking? One way is to add a new column of row numbers in each DataFrame. After adding this new column of row numbers, we can perform an inner join to get the required DataFrames vertically stacked.
We have to read two tables, called firsthorizontable and secondhorizontable, from the PostgreSQL database and create DataFrames.
We have to perform horizontal stacking of these newly created DataFrames.
How It Works
Note
In order to connect with the PostgreSQL database, we have to start a PySpark shell with the following options:
$pyspark --driver-class-path ~/.ivy2/jars/org.postgresql_postgresql-42.2.4.jar --packages org.postgresql:postgresql:42.2.4
Step 5-6-1. Reading Tables from PostgreSQL and Creating DataFrames
Step 5-6-2. Performing Horizontal Stacking of Newly Created DataFrames
And finally we have our required result.
Recipe 5-7. Perform Missing Value Imputation
Problem
You want to perform missing value imputation in a DataFrame.
Solution
For the data scientist, missing values are inevitable. PySparkSQL provides tools to handle missing data in DataFrames. Two important functions that deal with missing or null values are dropna() and fillna().

DataFrame with missing values
The dropna() function can remove rows that contain null data. It takes three arguments. The first argument is how, and it can take two values—any or all. If the value of how is all, a row will be dropped only if all the values of the row are null. If the value of how is any, the row will be dropped if any of its values are null.
The second argument of the dropna() function is called thresh. The default value for thresh is None. It takes an integer value. The thresh argument overwrites the first argument, how. If thresh is set to the integer n, all rows where the number of non-null values is less than n will be dropped.
The last argument of the dropna() function is subset. This is an optional name of columns to be considered.
The second important function for dealing with null values is fillna(). The first argument is a value. The fillna() function will replace any null value with the value argument .